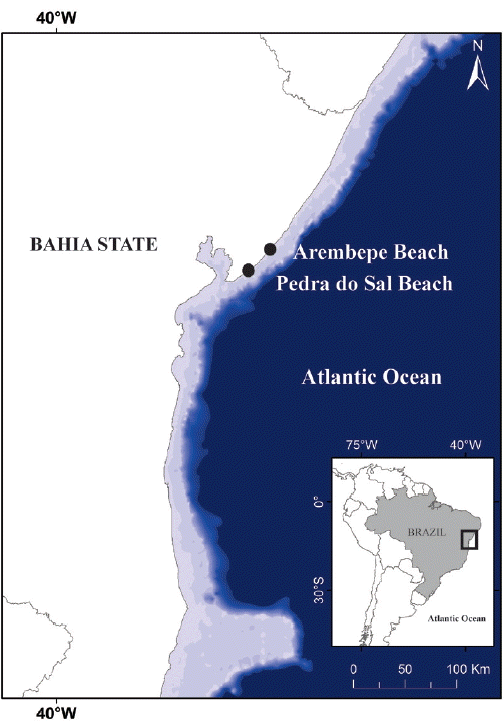
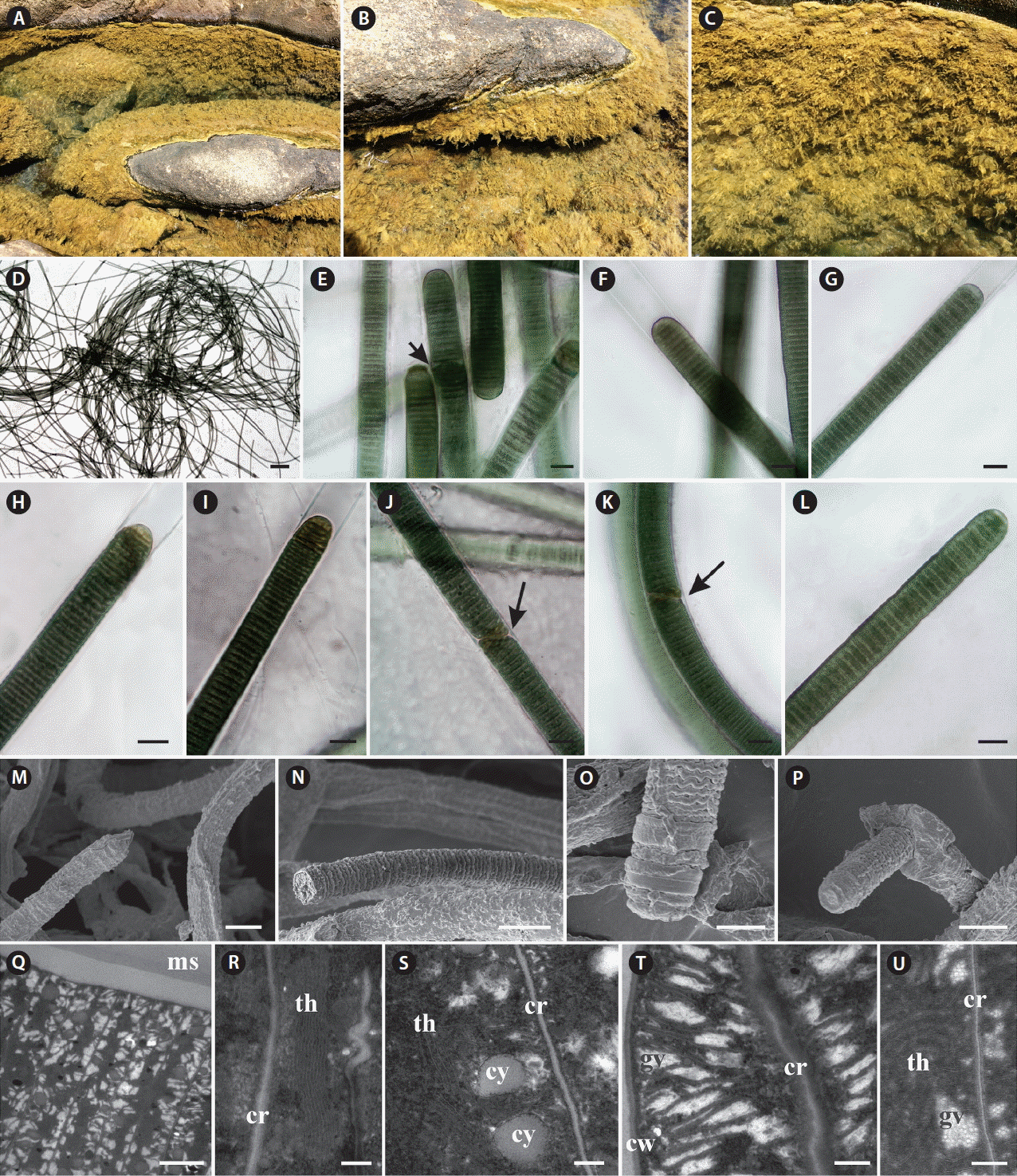
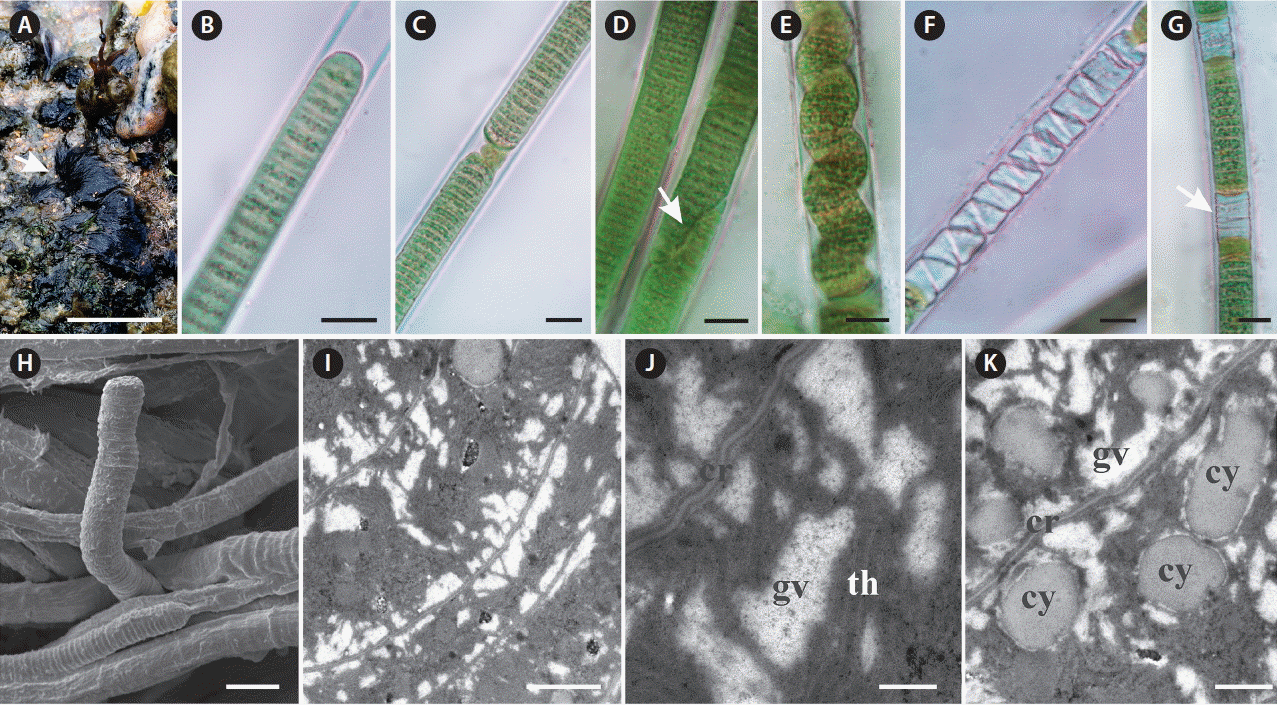
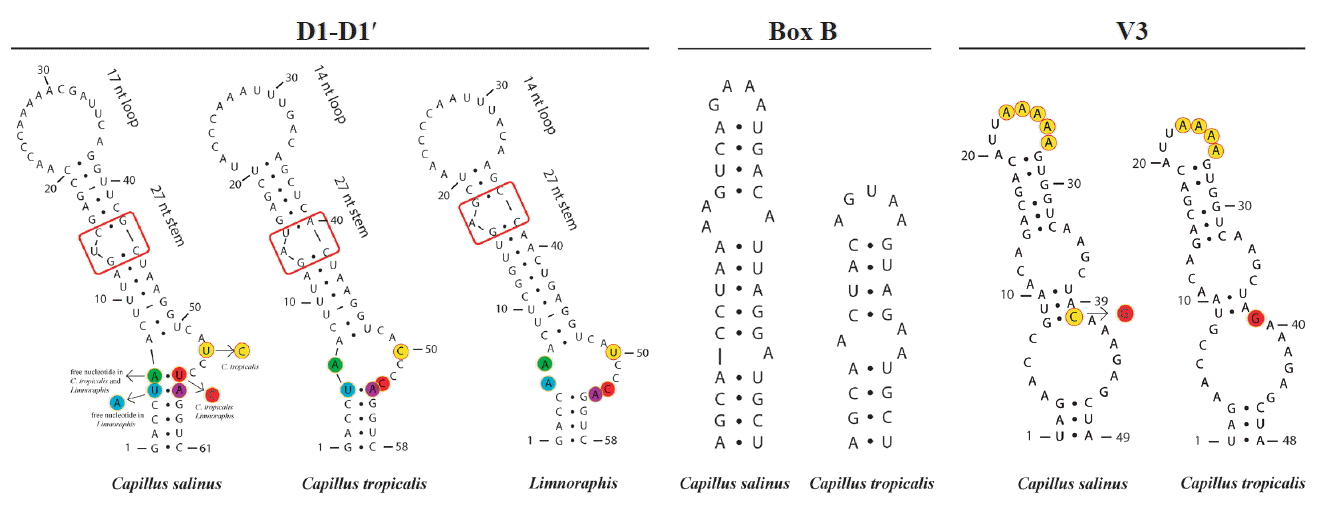
Baeta-Neves, MHC 1991. Estudo das Cianofíceas Marinhas Bentônicas da Região de Cabo Frio (Estado do Rio de Janeiro, Brasil). II–Hormogonae Rev Bras Biol. 52:641–659.
Boyer, SL, Flechtner, VR & Johansen, JR 2001. Is the 16S–23S rRNA internal transcribed spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Mol Biol Evol. 18:1057–1069.


Boyer, SL, Johansen, JR, Flechtner, VR & Howard, GL 2002. Phylogeny and genetic variance in terrestrial
Microcoleus (Cyanophyceae) species based on sequence analysis of the 16S rRNA gene and associated 16S–23S ITS region. J Phycol. 38:1222–1235.

Brito, A, Ramos, V, Mota, R, Lima, S, Santos, A, Vieira, J, Vieira, CP, Kaštovský, J, Vasconcelos, VM & Tamagnini, P 2017. Description of new genera and species of marine cyanobacteria from the Portuguese Atlantic coast. Mol Phylogenet Evol. 111:18–34.


Caires, TA, de Mattos Lyra, G, Hentschke, GS, de Gusmão Pedrini, A, Sant’Anna, CL & de Castro Nunes, JM 2018.
Neolyngbya gen. nov. (Cyanobacteria, Oscillatoriaceae): a new filamentous benthic marine taxon widely distributed along the Brazilian Coast. Mol Phylogenet Evol. 120:196–211.


Caires, TA, Sant’Anna, CL & de Castro Nunes, JMC 2013. A new species of marine benthic cyanobacteria from the infralittoral of Brazil: Symploca infralitoralis sp. nov. Braz. J Bot. 36:159–163.
Castenholz, RW, Rippka, R & Herdman, M 2001. Form-genus VII. Lyngbya Oxygenic photosynthetic bacteria. In : Boone DR, Castenholz RW, Garrity GM, editors Bergey’s Manual of Systematic Bacteriology. Springer, New York, 473–599.
Crispino, LMB & Sant’Anna, CL 2006. Cianobactérias marinhas bentônicas de ilhas costeiras do Estado de São Paulo, Brasil. Rev Bras Bot. 29:639–656.

Drummond, AJ, Ashton, B, Buxton, S, Cheung, M, Cooper, A, Duran, C, Field, M, Heled, J, Kearse, M, Markowitz, S, Moir, R, Stones-Havas, S, Sturrock, S, Thierer, T & Wilson, A 2011. Geneious v6.0.6, Available from:
http://www.geneious.com
. Accessed Oct 8, 2017
Engene, N, Choi, H, Esquenazi, E, Rottacker, EC, Ellisman, MH, Dorrestein, PC & Gerwick, WH 2011. Underestimated biodiversity as a major explanation for the perceived rich secondary metabolite capacity of the cyanobacterial genus
Lyngbya
. Environ Microbiol. 13:1601–1610.



Engene, N, Coates, RC & Gerwick, WH 2010. 16S rRNA gene heterogeneity in the filamentous marine cyanobacterial genus
Lyngbya
. J Phycol. 46:591–601.

Engene, N, Rottacker, EC, Kaštovský, J, Byrum, T, Choi, H, Ellisman, MH, Komárek, J & Gerwick, WH 2012.
Moorea producens gen. nov., sp. nov. and
Moorea bouillonii comb. nov., tropical marine cyanobacteria rich in bioactive secondary metabolites. Int J Syst Evol Microbiol. 62(Pt 5):1171–1178.



Fiore, MF, Moon, DH, Tsai, SM, Lee, H & Trevors, JT 2000. Miniprep DNA isolation from unicellular and filamentous cyanobacteria. J Microbiol Methods. 39:159–169.


Genuário, DB, Vaz, MGMV, Hentschke, GS, Sant’Anna, CL & Fiore, MF 2015.
Halotia gen. nov., a phylogenetically and physiologically coherent cyanobacterial genus isolated from marine coastal environments. Intern J Syst Evol Microbiol. 65:663–675.

Gomont, M 1892. Monographie des Oscillariées (Nostocacées Homocystées). Deuxième partie. - Lyngbyées
. Ann Sci Nat Bot Sér 7. 16:91–264.
Hentschke, GS, Johansen, JR, Pietrasiak, N, Fiore, MF, Rigonato, J, Sant’Anna, CL & Komárek, J 2016. Phylogenetic placement of
Dapisostemon gen. nov. and
Streptostemon, two tropical heterocytous genera (Cyanobacteria). Phytotaxa. 245:129–143.

Hentschke, GS, Johansen, JR, Pietrasiak, N, Rigonato, J, Fiore, MF & Sant’Anna, CL 2017.
Komarekiella atlantica gen. et sp. nov. (Nostocaceae, Cyanobacteria): a new subaerial taxon from the Atlantic Rainforest and Kauai, Hawaii. Fottea. 17:178–190.

Iteman, I, Rippka, R, Tandeau de Marsac, N & Herdman, M 2002. rDNA analyses of planktonic heterocystous cyanobacteria, including members of the genera
Anabaenopsis and
Cyanospira
. Microbiology. 148(Pt 2):481–496.


Jacinavicius, FR, Gama, WA Jr, Azevedo, MTP & Sant’Anna, CL 2012. Manual para cultivo de cianobactérias. Publicações Online do Instituto de Botânica de São Paulo, São Paulo, 32 pp.
Johansen, JR, Kovacik, L, Casamatta, DA, Fučiková, K & Kaštovský, J 2011. Utility of 16S–23S ITS sequence and secondary structure for recognition of intrageneric and intergeneric limits within cyanobacterial taxa:
Leptolyngbya corticola sp. nov. (Pseudanabaenaceae, Cyanobacteria). Nova Hedwigia. 92:283–302.

Karnovsky, MJ 1965. A formaldehyde-glutaraldehyde fixative of high osmolarity for use in electron microscopy. J Cell Biol. 27:1A–149A.

Komárek, J 2016. A polyphasic approach for the taxonomy of cyanobacteria: principles and applications. Eur J Phycol. 51:346–353.

Komárek, J 2018. Delimitation of the family Oscillatoriaceae (Cyanobacteria) according to the modern polyphasic approach (introductory review). Braz J Bot. 41:449–456.

Komárek, J & Anagnostidis, K 2005. Cyanoprokaryota-2 Teil/2nd part: Oscillatoriales. In : Büdel B, Gärtner G, Krienitz L, Schagerl M, editors Süsswasserflora von Mitteleuropa 19⁄778 2. Elsevier/Spektrum, Heidelberg, 1–759.
Komárek, J, Kaštovský, J, Mareš, J & Johansen, JR 2014. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia. 86:295–335.
Komárek, J, Zapomělová, E, Šmarda, J, Kopecký, J, Rejmánková, E, Woodhouse, J, Neilan, BA & Komárková, J 2013. Polyphasic evaluation of
Limnoraphis robusta, a water-bloom forming cyanobacterium from Lake Atitlán, Guatemala, with a description of
Limnoraphis gen. nov. Fottea. 13:39–52.

Lane, DJ 1991. 16S/23S rRNA sequencing. In : Stackebrandt E, Goodfellow M, editors Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons, Chichester, 115–175.
Lipkin, Y & Silva, PC 2002. Marine algae and seagrasses of the Dahlak Archipelago, southern Red Sea. Nova Hedwigia. 75:1–90.

Martins, MD & Branco, LHZ 2016.
Potamolinea gen. nov. (Oscillatoriales, Cyanobacteria): a phylogenetically and ecologically coherent cyanobacterial genus. Int J Syst Evol Microbiol. 66:3632–3641.


Mau, B, Newton, MA & Larget, B 1999. Bayesian phylogenetic inference via Markov chain Monte Carlo methods. Biometrics. 55:1–12.


McGregor, GB & Sendall, BC 2014. Phylogeny and toxicology of
Lyngbya wollei (Cyanobacteria, Oscillatoriales) from north-eastern Australia, with a description of
Microseira gen. nov. J Phycol. 51:109–119.


Mühlsteinová, R, Johansen, JR, Pietrasiak, N, Martin, MP, Osorio-Santos, K & Warren, SD 2014. Polyphasic characterization of
Trichocoleus desertorum sp. nov. (Pseudanabaenales, Cyanobacteria) from desert soils and phylogenetic placement of the genus
Trichocoleus
. Phytotaxa. 163:241–261.

Mukherjee, C, Chowdhury, R, Sutradhar, T, Begam, M, Ghosh, SM, Basak, SK & Ray, K 2015. Parboiled rice effluent: a wastewater niche for microalgae and cyanobacteria with growth coupled to comprehensive remediation and phosphorus biofertilization. Algal Res. 19:225–236.

Neilan, BA, Jacobs, D, Del Dot, T, Blackall, LL, Hawkins, PR, Cox, PT & Goodman, AE 1997. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus
Microcystis
. Int J Syst Bacteriol. 47:693–697.


Nylander, JAA 2008. MrModeltest 23 Program distributed by the author. Evolutionary Biology Centre. Uppsala University.
Porta, D & Hernández-Mariné, M 2005. Structural and ultrastructural characterization of several culture strains assigned to
Oscillatoria and
Lyngbya (Cyanophyta/Cyanoprokaryota/Cyanobacteria). Algol Stud. 117:349–370.

Posada, D & Buckley, TR 2004. Model selection and model averaging in phylogenetics: analysis advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53:793–808.


Reháková, K, Johansen, JR, Casamatta, DA, Xuesong, L & Vincent, J 2007. Morphological and molecular characterization of selected desert soil cyanobacteria: three species new to science including
Mojavia pulchra gen. et sp. nov. Phycologia. 46:481–502.

Rippka, R, Deruelles, J, Waterbury, JB, Herdman, M & Stanier, RY 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 111:1–61.

Ronquist, F, Teslenko, M, van der Mark, P, Ayres, DL, Darling, A, Höhna, S, Larget, B, Liu, L, Suchard, MA & Huelsenbeck, J 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.



Sambrook, JF & Russell, DW 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, 2100 pp.
Sant’Anna, CL 1995. Cyanophyceae marinhas bentônicas do Parque Estadual da Ilha do Cardoso, SP, Brasil. Hoehnea. 22:197–216.
Sant’Anna, CL 1997. Cyanophyceae marinhas bentônicas da região de Ubatuba, SP, Brasil. Hoehnea. 24:57–74.
Sant’Anna, CL, Cordeiro-Marino, M, Braga, MRA & Guimarães, SMPB 1985. Cianofíceas marinhas bentônicas das Praias de Peruíbe e dos Sonhos, Município de Itanhaém, São Paulo, Brasil. Rickia. 12:89–112.
Sciuto, K & Moro, I 2016. Detection of the new cosmopolitan genus
Thermoleptolyngbya (Cyanobacteria, Leptolyngbyaceae) using the 16S rRNA gene and 16S–23S ITS region. Mol Phylogenet Evol. 105:15–35.


Sharp, K, Arthur, KE, Gu, L, Ross, C, Harrison, G, Gunasekera, SP, Meickle, T, Matthew, S, Luesch, H, Thacker, RW, Sherman, DH & Paul, VJ 2009. Phylogenetic and chemical diversity of three chemotypes of bloom-forming
Lyngbya species (Cyanobacteria: Oscillatoriales) from reefs of southeastern Florida. Appl Environ Microbiol. 75:2879–2888.



Stamatakis, A 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.


Stamatakis, A, Hoover, P & Rougemont, J 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.


Vaz, MGMV, Genuário, DB, Andreote, APD, Malone, CFS, Sant’Anna, CL, Barbiero, L & Fiore, MF 2014.
Pantanalinema gen. nov. and
Alkalinema gen. nov.: novel pseudanabaenacean genera (Cyanobacteria) isolated from saline–alkaline lakes. Int J Syst Evol Microbiol. 65:298–308.


Werner, VR, Cabezudo, MM, Neuhaus, EB, Caires, TA, Sant’Anna, CL, Azevedo, MTP, Malone, C, Gama, WA Jr, Santos, KRS & Menezes, M 2017. Cyanophyceae in Lista de Espécies da Flora do Brasil. Jardim Botânico do Rio de Janeiro, Available from:
http://floradobrasil.jbrj.gov.br/jabot/floradobrasil/FB108025
. Accessed Aug 27, 2017