INTRODUCTION
Antarctica is an important location for exploring living organisms adapted to its harsh environment with relatively low anthropogenic impact. However, limited accessibility to Antarctica makes it difficult to collect samples and data. Nevertheless, several studies have examined Antarctic terrestrial life (Wall 2005, Convey et al. 2011, Dennis et al. 2019). The South Shetland Islands have diverse environments and valuable biological resources and are located 120 km north of the Antarctic Peninsula. Although several studies examined the diversity, ecology, and distribution of their terrestrial microalgae, taxonomic information on Antarctic microalgae remains limited (Fermani et al. 2007, Llames and Vinocur 2007, Zidarova 2008, Hamsher et al. 2016).
Members of the genus Micractinium Fresenius (family Chlorellaceae) are characterized by spherical or oval cells mostly arranged in colonies (Krienitz et al. 2004). The shape, size, and arrangement of cells and bristles are considered species-specific characters. However, the type species of the genus, Micractinium pusillum growing in dense culture as solitary spherical cells without bristles, is surprisingly morphologically similar to Chlorella vulgaris. Its morphological traits return to the original form while exposed to grazing zooplankton (Luo et al. 2005, 2006). This morphological similarity between the two genera causes difficulties when identifying taxa and defining them using morphology alone.
Recent phylogenetic studies of the secondary structure of internal transcribed spacer 2 (ITS2) have demonstrated characteristic differences among species with the typical morphology of Micractinium (Pröschold et al. 2010, 2011, Hoshina and Fujiwara 2013). The secondary structure of ITS2 differentiates species of Micractinium and Chlorella (Luo et al. 2010). Compensatory base changes (CBCs) and hemi-CBCs in the secondary structure of ITS2 can be useful for distinguishing individual species (Krienitz et al. 2004, 2010, Müller et al. 2007).
Because of their morphological simplicity and phenotypic plasticity, some algal collections tend to keep unclassified Antarctic coccoid green microalgal strains at species level. We investigated morphological and molecular characteristics of six Antarctic microalgal strains from the Korea Polar Research Institute (KOPRI) Culture Collections for Polar Microorganisms (KCCPM) in order to define morphologically ambiguous specimens at the species level. We have characterized three new species.
MATERIALS AND METHODS
Collections
Freshwater samples were collected from five sites on the South Shetland Islands, Antarctica in January 2014 (Table 1). Strains were isolated using sterile Pasteur pipettes. The cells were rinsed repeatedly and grown in artificial freshwater solution B5282 (Sigma-Aldrich, Saint Louis, MO, USA). They were cultivated at approximately 2°C under 25 μmol photons m−2 s−1 (16 : 8 h light : dark cycle) in a culture room at KCCPM.
Microscopy
Morphological identification was performed using an Axio Imager A2 (Carl Zeiss Inc., Hallbergmoos, Germany) equipped with differential interference contrast optics. Photomicrographs were taken with an AxioCam HRC camera (Carl Zeiss Inc.).
DNA extraction, amplification, and sequencing
DNA was extracted using the i-genomic Plant DNA Extraction Kit (iNtRON Biotechnology, Seongnam, Korea) according to the manufacturer’s instructions. We performed polymerase chain reaction (PCR) to amplify small subunit (SSU) rDNA, internal transcribed spacer 1 (ITS1), 5.8S rDNA, and ITS2 with the primer pairs NS1/NS4, NS5/NS8, and ITS1/ITS4 (White et al. 1990). The PCR products were purified with the MG PCR Product Purification Kit (Macrogen, Seoul, Korea) following the manufacturer’s recommendations. The DNA was sequenced by Macrogen.
Phylogenetic analyses
Phylogenetic analyses were performed on a concatenated dataset of the SSU, ITS1, 5.8S, and ITS2 rDNA sequences. The aligned sequences were checked for possible misaligned positions in BioEdit 7.0.5.3 (Hall 1999). The alignment was constructed by taking sequences of Micractinium species from previous studies (Krienitz et al. 2010, Luo et al. 2010, Hoshina and Fujiwara 2013) and comparing them to the newly obtained ones.
Appropriate evolutionary model was assessed using jModelTest 2 (Darriba et al. 2012) under the Akaike Information Criterion. Selected model has the following parameters: GTR + I + G with gamma distribution = 0.5220; proportion of invariable sites = 0.6780; base frequencies A = 0.2070, C = 0.2843, G = 0.2725, T = 0.2362; and rate among sites [A-C] = 0.8495, [A-G] = 1.1688, [A-T] = 1.1254, [C-G] = 0.6076, [C-T] = 3.1932, and [G-T] = 1.0000. Maximum likelihood (ML) analyses were conducted using PhyML ver. 3.0 (Guindon et al. 2010), applying the chosen evolutionary model with bootstrap analyses using 1,000 replicates. Bayesian inference (BI) was run using MrBayes 3.2.6 (Ronquist et al. 2012) with two simultaneous runs (nruns = 2) and four Metropolis-coupled Markov chain Monte Carlo (MC3) algorithms for 10 × 106 generations. The first 25% of the trees were discarded as burn-in. The trees were visualized using FigTree v.1.4.2 (available at http://tree.bio.ed.ac.uk/software/figtree/).
ITS2 secondary structure
To locate hemi-CBCs and CBCs, the ITS2 secondary structures were constructed using Mfold (Zuker 2003) and 4SALE (Seibel et al. 2006, 2008).
RESULTS
Morphological observations by light microscopy
The Antarctic strains KSF0085, KSF0094, KSF0100, KSF0112, KSF0114, and KSF0127 possessed the typical morphological characteristics of Micractinium species in terms of cell shape, chloroplasts, and pyrenoids (Table 2). However, bristles and colony formation were observed only for KSF0085. The vegetative cells of KSF0085 were spherical or ellipsoidal, and 5.0–5.4 × 8.2–8.6 μm in diameter with or without bristles in culture conditions (Fig. 1A & B). The cells contained a cup- to girdle-shaped chloroplast with a spherical to ellipsoidal pyrenoid. The cells of KSF0094 were spherical or ellipsoidal, and 4.5–4.7 × 7.2–7.4 μm in diameter (Fig. 1C). A single cup- or girdle-shaped chloroplast with a pyrenoid was present. KSF0112 was spherical to ellipsoidal, 3.3–3.9 × 5.5–5.7 μm, and contained cup- or girdle-shaped chloroplasts with a pronounced pyrenoid (Fig. 1D). KSF0100, KSF0114, and KSF0127 were morphologically similar to KSF0112.
Sequencing results and phylogenetic analyses
SSU sequence-based approach of the members of Chlorellaceae, did not provide enough information to distinguish taxa from public database as well as from this study at species level. Further analyses were performed using the combined SSU and ITS rDNA dataset. The genus Micractinium was highly supported in the ML and BI analyses (Fig. 2). The tree revealed 10 distinct lineages within the genus, including four lineages known as Micractinium species: M. inermum, M. conductrix, M. pusillum, and M. belenophorum. The six Antarctic strains (KSF0085, KSF0094, KSF0100, KSF0112, KSF0114, and KSF0127) clustered with non-bristle forming M. inermum in both analyses. The six strains were subdivided into two clades; one clade contained four strains (KSF0100, KSF0112, KSF0114, and KSF0127), and the other two strains formed two subclades. The strains (KSF0100, KSF0112, KSF0114, and KSF0127) formed first clade were collected from four sites of the South Shetland Islands and were identical to Micractinium sp. (KNUA029) (Hong et al. 2015). KSF0085 belonged to the same clade as Micractinium sp. (KNUA034), which is an Antarctic strain without bristles (Hong et al. 2015). KSF0085 was sister to KSF0094 with strong to moderate support in the analyses.
ITS2 secondary structure
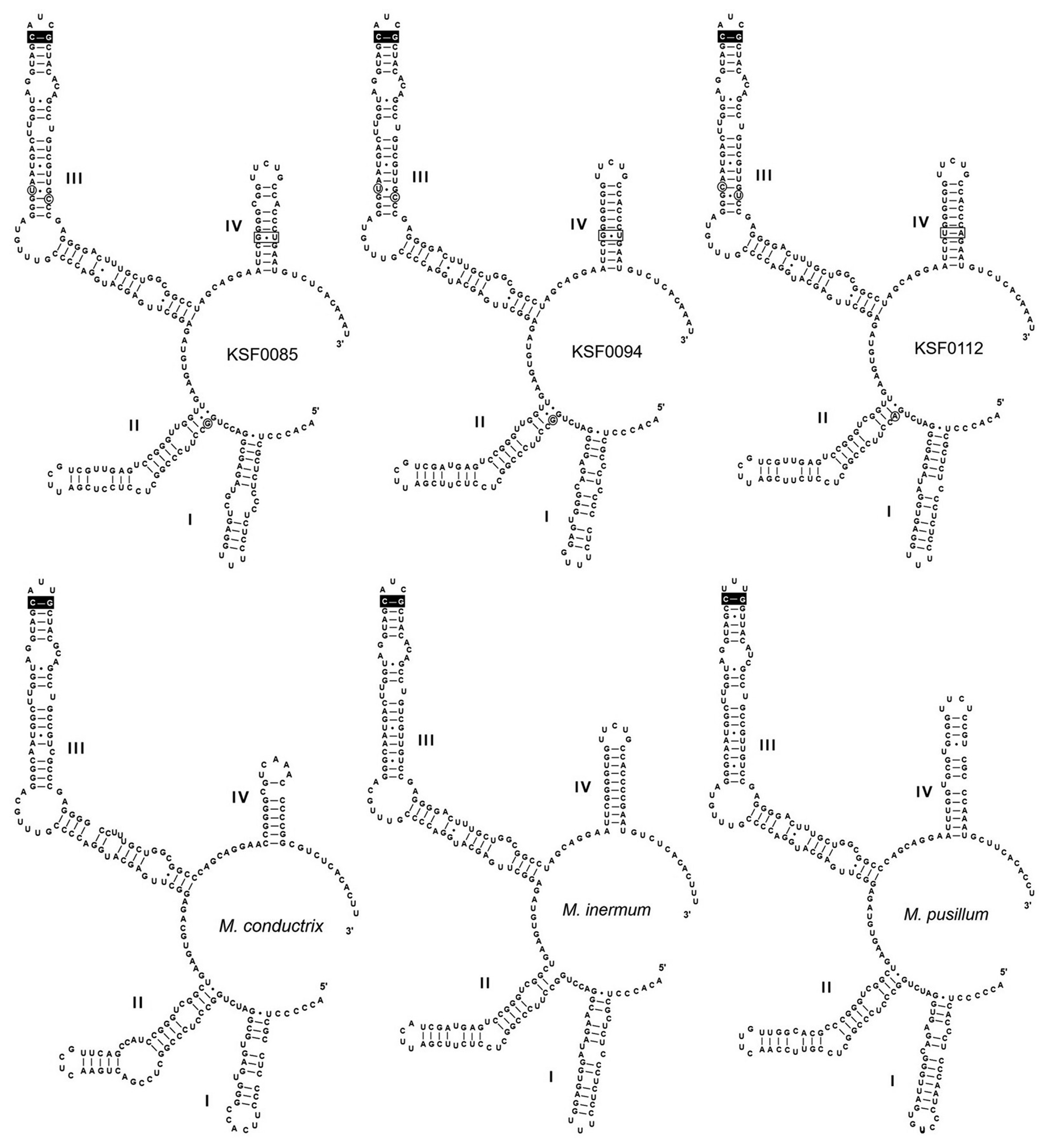
Fig. 3 shows the secondary structures of ITS2 of the Antarctic strains and other species of Micractinium. All structures possess motifs conserved within the green algae (Mai and Coleman 1997), including a four-fingered hand (helices I-IV), pyrimidine–pyrimidine bulge on helix II, and a conserved sequence TGGT (UGGU) on the 5′-side of helix III. Micractinium species have C-G pairing at the tip of helix III (Luo et al. 2006, 2010). We found distinct characteristics in the ITS2 secondary structures of the six Antarctic strains, with three types of ITS2 secondary structure (Fig. 3). These structures and base changes differed from the closely related species M. inermum. Compared to those of M. inermum, four hemi-CBC sites of ITS2 secondary structures were revealed in KSF0085 and KSF0094, and two CBC sites were revealed in KSF0112. KSF0085 and KSF0094 showed differences in helix I structure, and no CBC or hemi-CBC was found in these strains. KSF0112 showed a base change from G : U to U : A in helix IV and three hemi-CBCs in helices II and III, compared to KSF0085 and KSF0094.
DISCUSSION
The family Chlorellaceae, containing small, round, non-flagellate microalgae, has been divided into several groups using various characteristics, such as colony formation, bristle, mucilage, and pyrenoid (Krienitz et al. 2004). They recognized Micractinium has spherical to ellipsoidal cells with bristle formation in a colony. However, not all Micractinium species are capable of bristle producing (Luo et al. 2010, Hoshina and Fujiwara 2013), and the differentiation of Micractinium species based on the length and number of bristles is unreliable (Luo et al. 2006). Luo et al. (2005) observed single-celled and bristle-free forms when M. pusillum, the type species of the genus, was cultivated without zooplankton. However, culturing Micractinium strain with zooplankton does not always produce bristles and colonies (Pröschold et al. 2011). M. conductrix, M. inermum, and other Antarctic strains (KNUA029 and KNUA034) of the genus are non-bristle and similar to Chlorella morphologically (Pröschold et al. 2011, Hoshina and Fujiwara 2013, Hong et al. 2015). Species delimitation in Micractinium has always been problematic because there are few diagnostic morphological characters and they are not easily observed. Just one out of six Antarctic strains, KSF0085 produced various colony types and bristle formations when observed in the light microscope, whereas the other strains did not show these typical characteristics of Micractinium (Fig. 1).
Phylogenetic analyses of the SSU and ITS sequences of the nuclear rDNA gene revealed that the Antarctic Micractinium strains were clearly separated from the genus Chlorella and these strains were closely related to M. inermum (Fig. 2). Hong et al. (2015) reported that KNUA029 and KNUA034 were collected from two sites in West Antarctica. Morphological and molecular characteristics of KNUA029 are identical with four Antarctic strains (KSF0100, KSF0112, KSF0114, and KSF0127) and those of KNUA034 are same with strain KSF0085.
Recent molecular studies have demonstrated the usefulness of ITS2 sequences for defining species of green coccoid algae. The ITS2 region is highly conserved within species (Müller et al. 2005), whereas it is highly divergent between species (Hoshina et al. 2010). This strongly encourages a species concept based on ITS2 sequence differences (Mai and Coleman 1997). In addition, many taxonomic studies have used CBC in the ITS2 secondary structure for species delimitation (Krienitz et al. 2004, Müller et al. 2007, Hoshina and Fujiwara 2013). The distinctive base pairing of helix III of ITS2 clearly distinguishes Micractinium from other genera within the Chlorella clade (Luo et al. 2010). As our study showed, the six investigated strains belong to the genus Micractinium based on the C-G pairing CBC at the tip of ITS2 helix III (Fig. 3). The six Antarctic strains are considered to be three separate species, with differences in the secondary structure of ITS2, CBCs, and hemi-CBCs. KSF0085 and KSF0094 showed the greatest difference in helix I structure among the ITS2 secondary structures in the Antarctic strains. KSF0112 showed differences in one CBC and three hemi-CBCs with KSF0085 and KSF0094. The morphological and molecular characteristics of KSF0100, KSF0112, KSF0114, and KSF0127 were identical.
There are members of the genus Micractinium that are distributed worldwide, but no species has been reported yet in Antarctica (Guiry and Guiry 2019). The Antarctic strains show tolerance to lower temperature and produced polyunsaturated fatty acids in cold condition (Hong et al. 2015). It suggests an evolutionary evidence of their adaptation to the Antarctic environment. The six Antarctic strains are similar to other Micractinium species morphologically (Table 2). However, these Antarctic strains are clearly discriminated from other Micractinium species by molecular characteristics such as their phylogenetic relationships and ITS2 secondary structures, therefore we propose them as three new species.
Taxonomic conclusion
Micractinium simplicissimum H. Chae, H. -G. Choi & J. H. Kim sp. nov. (Fig. 1D)
Cells solitary, planktonic, spherical to ellipsoidal shaped, 3.3–3.9 × 5.5–5.7 μm, without bristles. Mucilage absent. Chloroplast single, parietal, cup-, girdle-shaped, with a broadly ellipsoidal to spherical pyrenoid. Reproduction by autospores, zoospores not observed. Differs from other species of this genus by the order of nucleotides in ITS2 and the barcoding signatures.
Holotype
KSF0112 collected on Jan 11, 2014 in King George Island (South Shetland Islands, Antarctica; 62°14′24.49″ S, 58°43′29.18″ W) by S. Lim and stored at KCCPM, Incheon, Korea.
Micractinium singularis H. Chae, H. -G. Choi & J. H. Kim sp. nov. (Fig. 1C)
Cells solitary, planktonic, spherical to ellipsoidal shaped, 4.5–4.7 × 7.2–7.4 μm, without bristles. Mucilage absent. Chloroplast single, parietal, cup-, girdle-shaped, with a broadly ellipsoidal to spherical pyrenoid. Reproduction by autospores, zoospores not observed. Differs from other species of this genus by the order of nucleotides in ITS2 and the barcoding signatures.
Micractinium variabile H. Chae, H. -G. Choi & J. H. Kim sp. nov. (Fig. 1A & B)
Cells solitary or colonial, planktonic, spherical to ellipsoidal shaped, 5.0–5.4 × 8.2–8.6 μm, with or without bristles. Mucilage absent. Chloroplast single, parietal, cup-, girdle-shaped, with a broadly ellipsoidal to spherical pyrenoid. Reproduction by autospores, zoospores not observed. Differs from other species of this genus by the order of nucleotides in ITS2 and the barcoding signatures.