INTRODUCTION
Massive blooms of green algae, generally referred to as green tides, have been increasing in frequency worldwide (Smetacek and Zingone 2013). Recurrent green tides caused by Ulva species are associated with eutrophication along enclosed coasts and have strong impacts on aquaculture, tourism, and marine ecosystems (Cui et al. 2018). The genus Ulva Linnaeus, which includes approximately 130 taxonomically accepted species (Guiry and Guiry 2019), is well known to form blooms and tolerate a wide range of salinities (Leliaert et al. 2009, Wan et al. 2017). Members of this genus are found in various habitats and environments, especially in coastal areas with high levels of eutrophication (Kang and Chung 2017), and can increase in biomass explosively due to their opportunistic features of rapid nutrient uptake and fast growth rate (Melton et al. 2016, Hu et al. 2017). The world’s largest green tide caused by Ulva prolifera has occurred in the Yellow Sea of China annually from 2007 to 2017, for 11 continuous years (Zhang et al. 2019). Previous reports of green tides in Japan have suggested that the major bloom-forming green alga was Ulva ohnoi, representing a unique ecotype separate from adjacent populations of Ulva spp. (Hiraoka et al. 2003, Kawai et al. 2007). Ulva blooms in the Yellow Sea rapidly generate a huge amount of biomass that can be transported to the western coast of the Korean Peninsula (Kang et al. 2014).
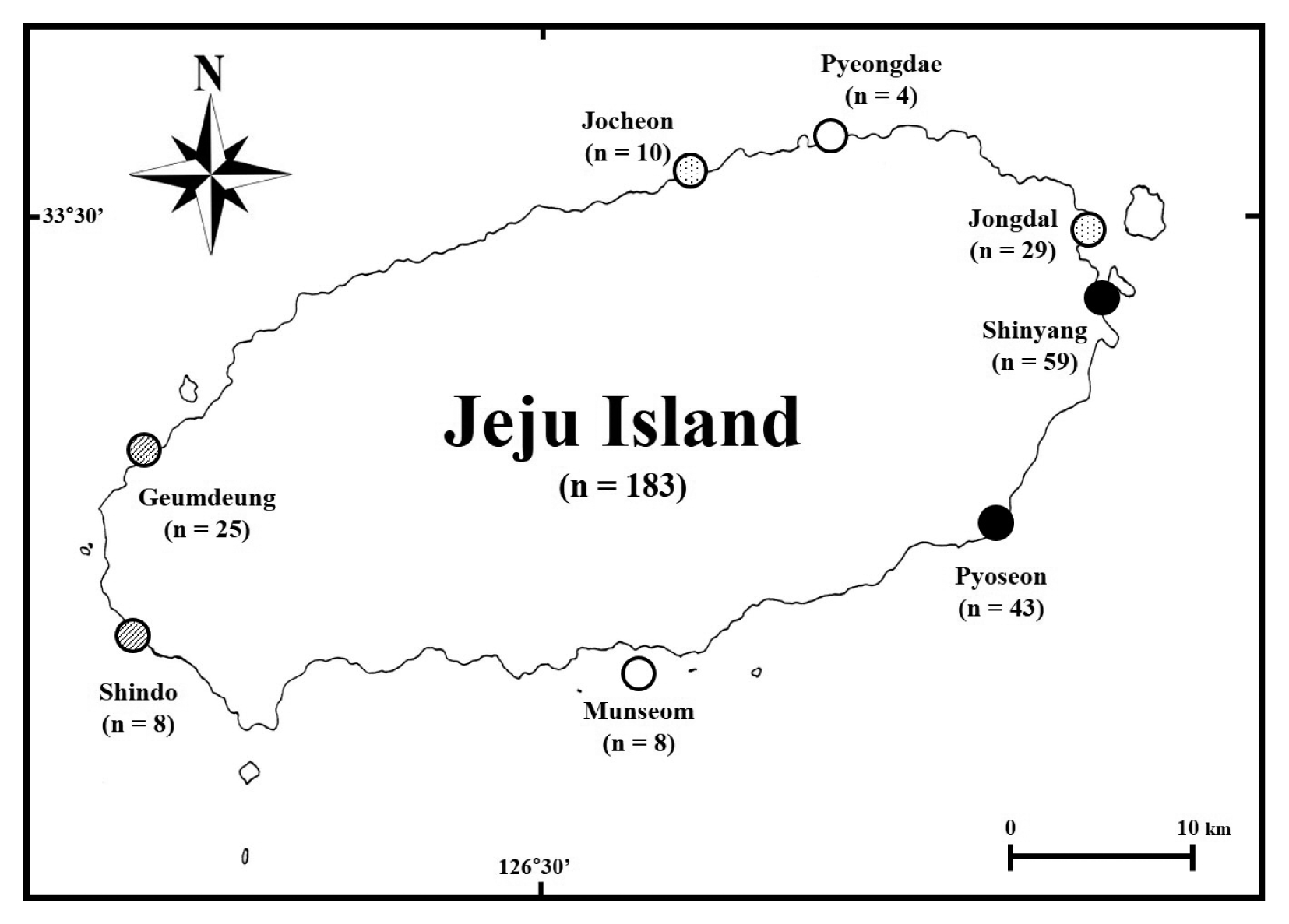
Green tides around Jeju Island usually start at the end of April, and last for approximately 6 months until the end of October, although some parts of the coast of Jeju Island maintain Ulva biomass throughout the year (personal communication). Green tides of Ulva have occurred along the eastern coast of Jeju Island almost every year since the early 2000s (Song et al. 2016). In particular, Shinyang is the most famous site of such blooms due to benthic eutrophication caused by anthropogenic and geological factors (Hwang et al. 2005, Song et al. 2016). Furthermore, bloom occurrence has increased with marine environmental changes, including those caused by global warming (Kwon et al. 2017). In addition, Jocheon harbor, located in the northern part of the island, has experienced increasing annual problematic blooms due to its semi-enclosed embayment and submarine groundwater discharge (Kwon et al. 2017).
Although definitive taxonomy is an essential cornerstone to understanding the occurrence and driving mechanisms of massive blooms of Ulva species in coastal areas (Leliaert et al. 2009), there are serious challenges to correctly identifying Ulva specimens based on habit and microscopic characteristics due to morphological plasticity along ecological and environmental gradients (Bermejo et al. 2019). Therefore, molecular genetic information is necessary to identify Ulva, and DNA barcoding is one method of obtaining such information. Hebert et al. (2003) proposed DNA barcoding based on short and standardized sequences for identification of eukaryotic organisms. Specifically, the plastid marker, elongation factor Tu (tufA), has been recommended as a standard marker for the routine barcoding of green marine macroalgae due to its strong amplification and lack of introns (Saunders and Kucera 2010).
The tufA barcoding marker has been applied to delimiting species boundaries of green macroalgal taxa including Caulerpa (Belton et al. 2014), Codium (Lee and Kim 2015), Halimeda (Cremen et al. 2016), and Ulva (Spalding et al. 2016). Although several studies have focused on the factors causing massive blooms around Jeju Island (Hwang et al. 2005, Song et al. 2016, Kwon et al. 2017), little attention has been paid to the taxonomic identification of the Ulva species involved in these blooms. Recently, a study of Ulva from Jeju Island was performed to clarify its species composition (Kang et al. 2019). They focused on the species diversity along the coast of Jeju without precisely determining the causative species of Ulva blooms. Our study, therefore, aims to reveal the major Ulva species that cause massive blooms around Jeju Island. To accomplish this goal, specimens of Ulva from eight sites were sampled on Jeju Island including the ecologically problematic blooming sites at Shinyang and Jocheon, and identified using tufA and ribulose-1,5-biphosphate carboxylase large subunit (rbcL) DNA barcoding sequences.
MATERIALS AND METHODS
A total of 183 Ulva samples were collected from eight sites (Supplementary Table S1) from August 2017 to March 2018. The timing of Ulva collection was random, as its species are known to regularly cause massive green tides around Jeju Island (Kwon et al. 2017). The Ulva specimens from the blooming sites were collected from intertidal to subtidal. Considering bloom level and the characteristics of the coastal environment (Table 1, Fig. 1), we selected eight collection sites, including two embayment sites (solid circles in Fig. 1), two harbors (dotted circles in Fig. 1), two intertidal zones with artificial structures such as a pool or dike (hatched circles in Fig. 1), and two open coast sites (empty circles in Fig. 1). Among the two embayment and two harbor sites, Shinyang (embayment) and Jocheon (harbor) both have reported massive annual blooms (Song et al. 2016, Kwon et al. 2017), while Pyoseon (embayment) and Jongdal (harbor) have similar costal environments and are sites of less serious blooms. Geumdeung and Shindo represent intertidal zone sites where blooms are possible.
The specimens were photographed using an Olympus TG-4 camera (Olympus, Tokyo, Japan) to assist in the description of their external habits. Dried samples for DNA barcoding were prepared using silica gel. Samples preserved with 10% formalin in seawater were sectioned using a bench-top freezing microtome (NK-101-II; Nippon Optical Works Co., Ltd., Tokyo, Japan) and photographed under a microscope (BX43; Olympus) using an EOS 600D digital camera (Canon, Tokyo, Japan) to characterize the vegetative structures. Digitized images were edited for clarity using Adobe Photoshop software (v. 6.1; Adobe Systems Inc., San Jose, CA, USA). Pressed herbarium sheets were deposited as voucher specimens in the herbarium of Jeju National University (JNUB), Korea.
Total genomic DNA was extracted following the protocol of the LaboPass Tissue Genomic DNA Isolation Kit Mini (Cosmo Gentech, Seoul, Korea). AccuPower PCR Premix (Bioneer, Daejeon, Korea) was used according to the manufacturer’s protocol for all PCR reactions. Two full-length plastid genes were amplified, tufA and the rbcL, to allow for species identification and phylogenetic analysis of Ulva specimens. The primer combinations used for tufA and rbcL were TufGF4 / TufGR (Saunders and Kucera 2010) and RH1 / 1385R (Manhart 1994), respectively. The PCR amplification procedure followed that of Saunders and Kucera (2010) for tufA and Heesch et al. (2009) for rbcL. All successfully amplified PCR products were purified using an AccuPrep PCR Purification Kit (Bioneer), and were sent to Macrogen (Seoul, Korea) for forward and reverse sequencing. All representative tufA and rbcL sequences from GenBank were selected for comparison: 25 in the tufA dataset; 44 for rbcL (Supplementary Table S2). Both sequence datasets were aligned visually using BioEdit (Hall 1999) after editing the Ulva sequences obtained in this study using Chromas ver. 1.45 software (Technelysium Pty Ltd., South Brisbane, Australia). We assessed the levels of variation in the tufA and rbcL sequences based on uncorrected pair-wise genetic distance (p-distance) estimates obtained from MEGA 5.1 software using a neighbor-joining algorithm dependent on the Kimura-2-parameter distance method. To construct the combined phylogenetic tree based on both tufA and rbcL sequence datasets, maximum likelihood (ML) analyses were performed using RAxML software (Stamatakis 2006) and the ML combined tree was edited with FigTree (v1.4.0). RAxML analysis was performed with all three codons partitioned under the GTR + Γ + I model using 1,000 bootstrap replicates. In addition, TCS 1.21 (Clement et al. 2000) was used to construct haplotype networks based on tufA and rbcL sequence data at the 95% confidence level.
RESULTS
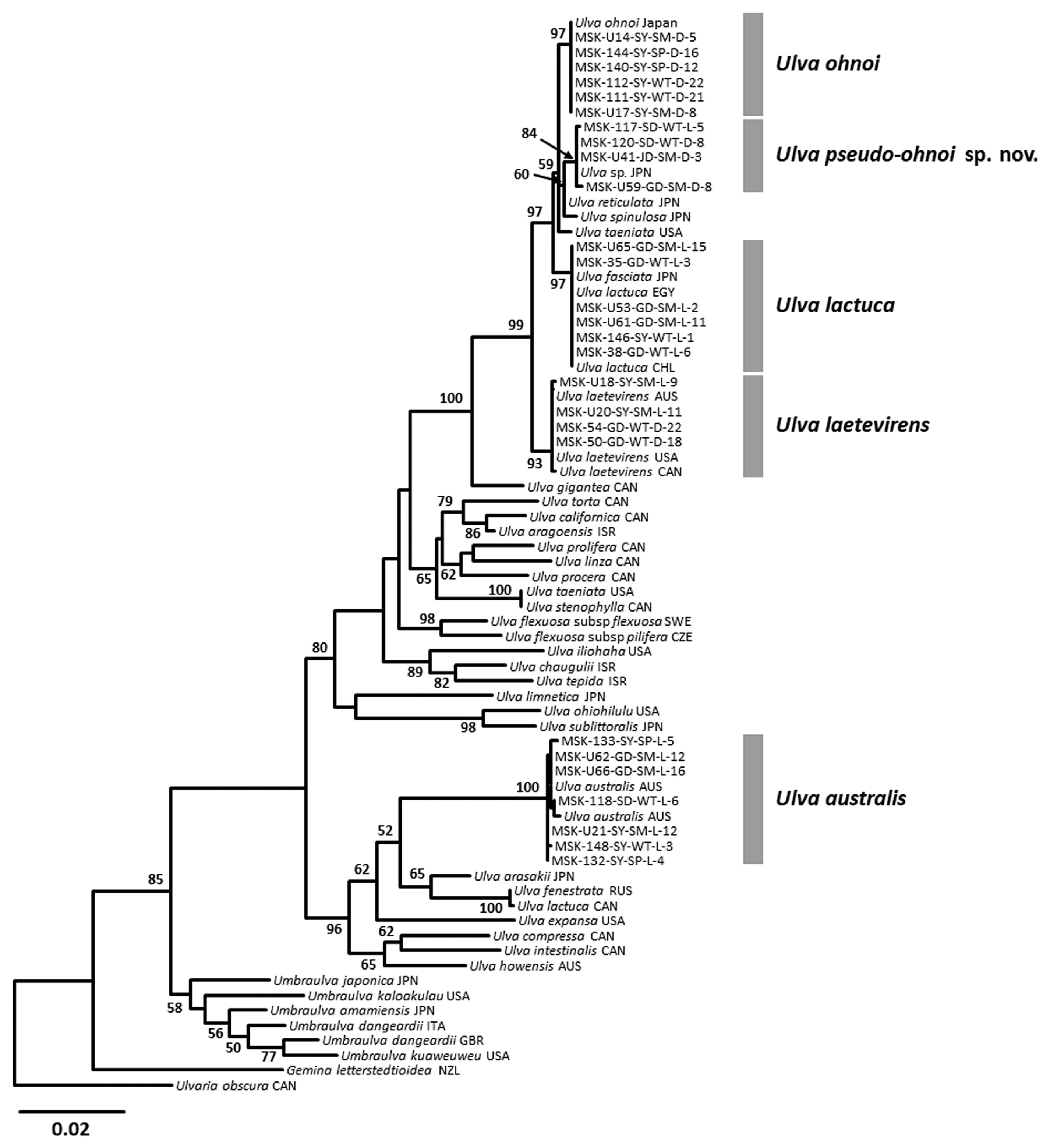
DNA barcodes were acquired for 172 Ulva bloom specimens on the basis of tufA ranged from 708–866 bp in length, and resolved into five genetic groups (Supplementary Table S1). These groups were assignable to Ulva australis Areschoug, Ulva lactuca Linnaeus, Ulva laetevirens Areschoug, Ulva ohnoi Hiraoka & Shimada, and a novel species-level group. The intraspecific variation was 0–0.4% within U. australis, 0–0.9% within U. laetevirens, and 0–0.1% within U. ohnoi and the novel group. No genetic variation was observed within U. lactuca. The interspecific variations ranged from 0.6–0.7% between U. ohnoi and the novel group to 1.5–2.5% between U. laetevirens and the novel group. The interspecific variation between U. australis and others was 10.7–12.4%. The rbcL results included 169 sequences of up to 1,364 bp (Supplementary Table S1), which exhibited the same five genetic groups with lower intraspecific variations compared than tufA. The intraspecific variation was 0–0.2% within U. australis, 0–0.3% within U. ohnoi, and 0–0.1% within U. lactuca, U. laetevirens and the novel group. The interspecific variation was 0.4–0.8% among four genetic groups except U. australis. U. australis showed 2.8–3.3% interspecific variations to four other groups.
Combined ML analysis of tufA and rbcL was performed to define the taxonomic positions and resolve phylogenetic relationships among U. australis, U. laetevirens, U. lactuca, U. ohnoi, and the novel species (Fig. 2). Each of the four species and the novel species-level group were strongly supported in the combined tufA and rbcL phylogeny, with bootstrap values ranging from 84% for the novel group, to 100% for U. australis. U. australis diverged early from the other four specific groups, and those four groups formed a monophyletic clade. The novel group was sister to U. ohnoi with moderate bootstrap support of 69% based on tufA, and diverged as a sister group to U. lactuca and U. ohnoi supported with 73% bootstrap support based on rbcL. In the phylogenetic analysis using a concatenated matrix of tufA and rbcL sequences, the novel species was more clearly divergent from the clade of U. lactuca and U. ohnoi, with good bootstrap support (84%) (Fig. 2). Based on these phylogenetic results, we propose the new species, U. pseudo-ohnoi sp. nov.
The four species appeared at the embayment site of Shinyang, where the most severe blooms occur (Table 1). Among the 59 specimens analyzed based on tufA and rbcL, U. ohnoi was dominant, comprising 33 specimens. Although another embayment site, Pyoseon, also hosted four species and was identical to the species composition from Shinyang, U. ohnoi was not dominant like at Shinyang (Table 1). In Pyoseon, the three species, U. australis, U. lactuca, and U. ohnoi, occurred equally. At the two harbor sites, three species were observed in equal amounts, these were U. australis, U. laetevirens, and the novel species (Table 1). At Jocheon harbor, the novel species was dominant, while U. laetevirens was more common at Jongdal harbor (Table 1). At Geumdeung, where huge tide pools characterize the habitat, four species were present, including U. australis, U. lactuca, U. laetevirens, and the novel species (Table 1). At Shindo, which has an artificial dike structure, U. australis and the novel species occurred (Table 1). Pyeongdae and Munseom are open coastal areas with less frequent blooms, and each hosted one species each with a distromatic and free-floating habit, U. australis and U. lactuca, respectively (Table 1).
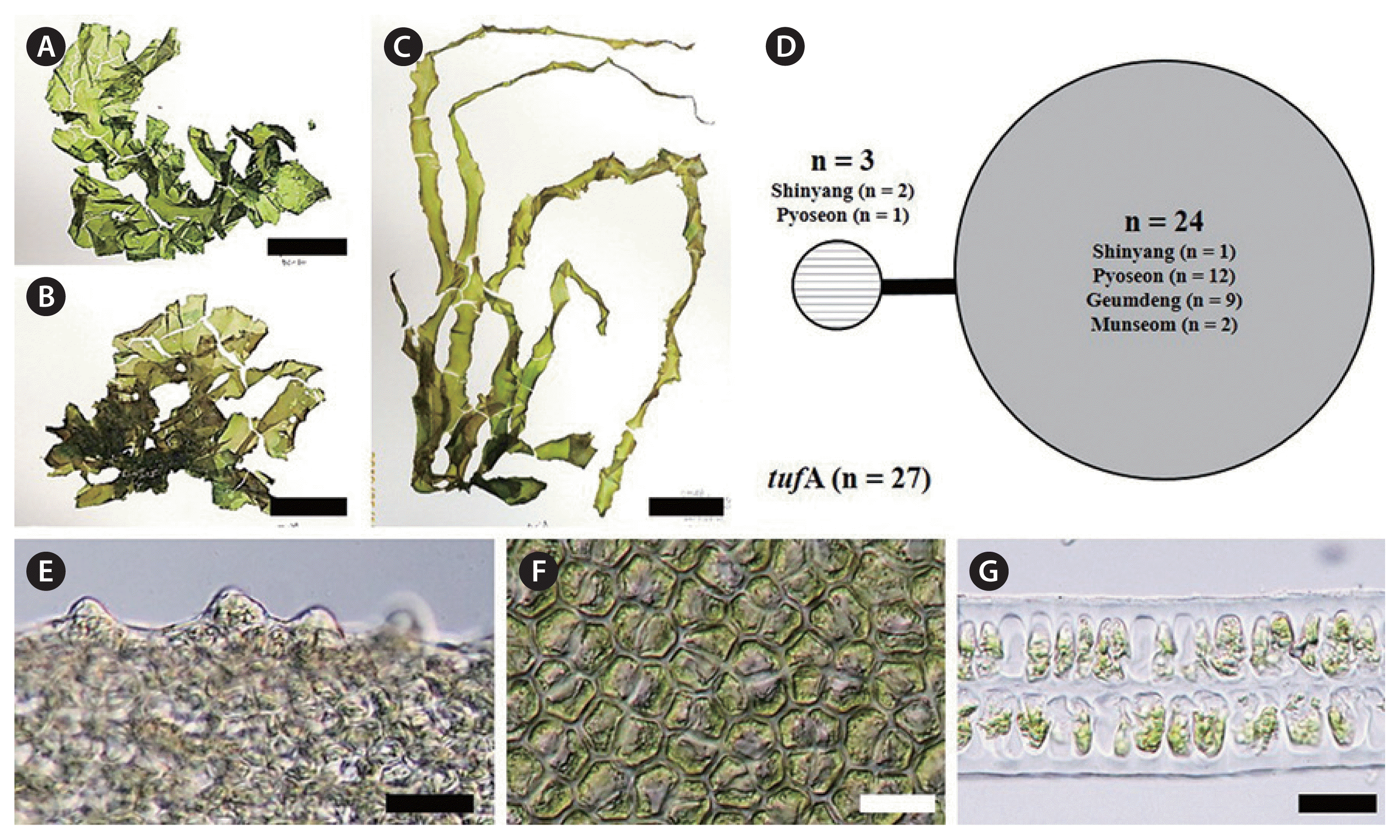
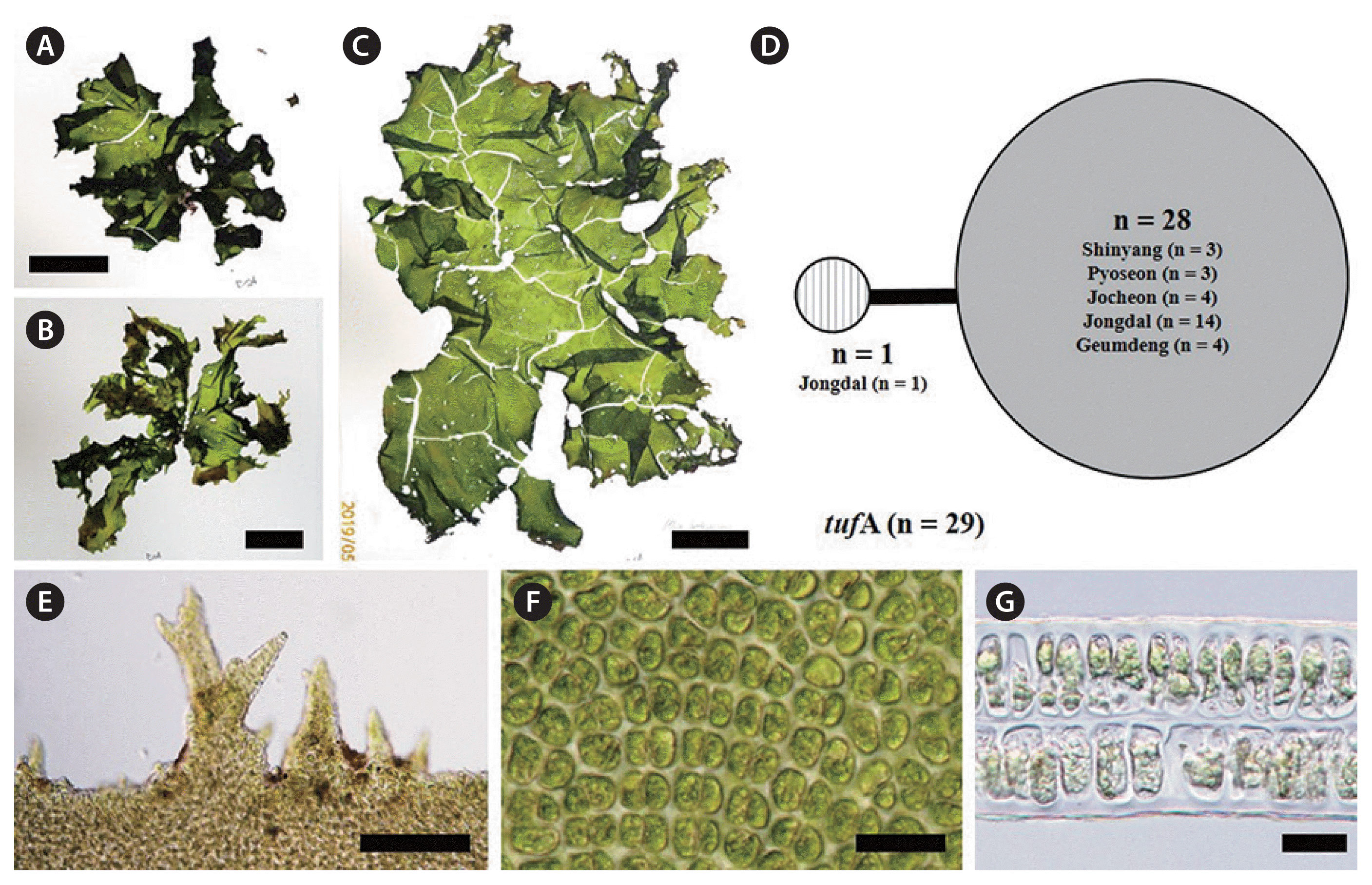
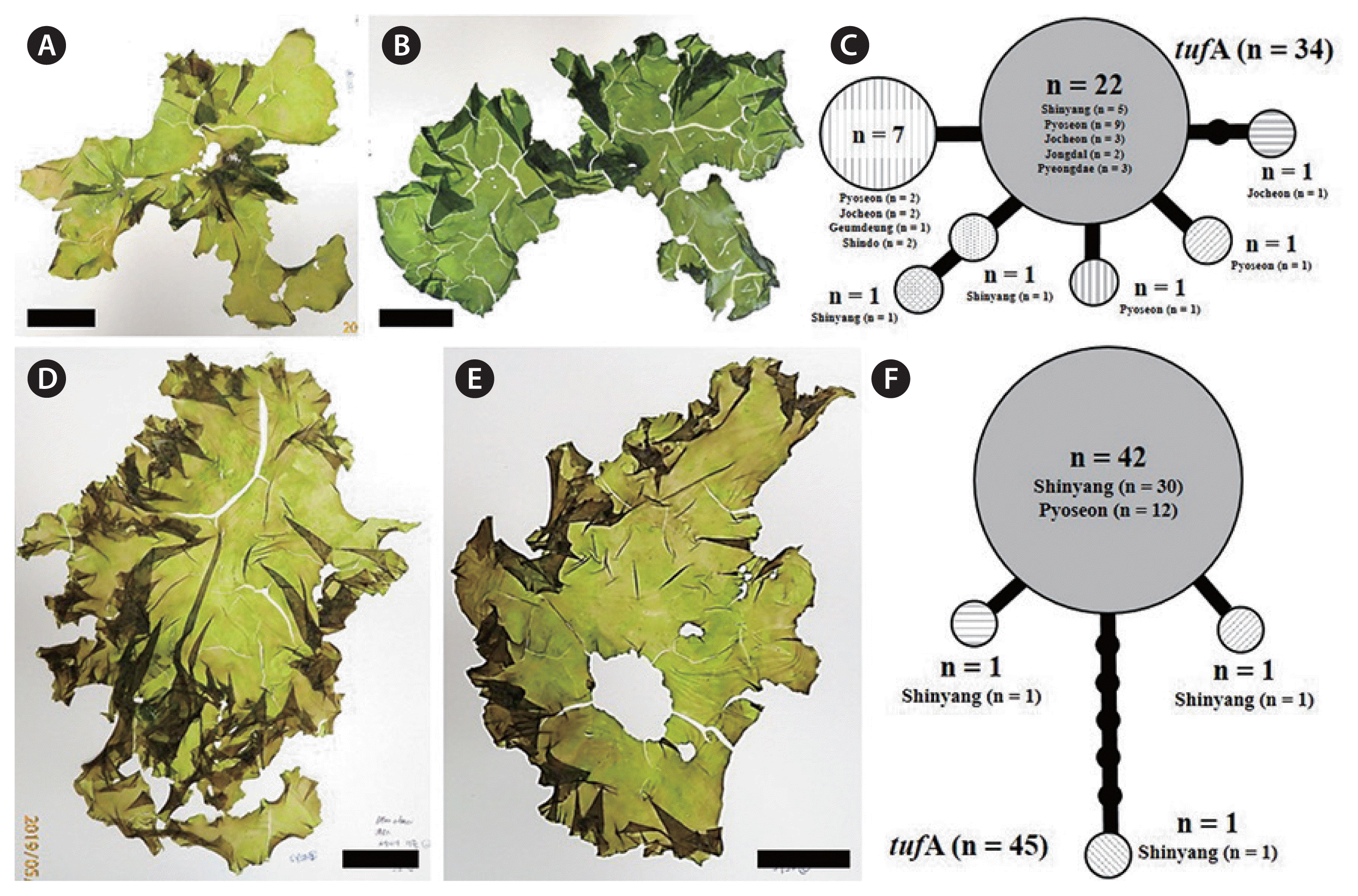
The haplotype networks of five species from bloom sites around Jeju Island were low levels of genetic diversity (Figs 3C, 4D, 5D, 6C & F). Examination of the tufA networks indicated that the novel species (Fig. 3C), U. lactuca (Fig. 4D), and U. laetevirens (Fig. 5D) were each composed of only two haplotypes. U. australis showed the greatest variation (Fig. 6C) including the center (n = 22) containing samples from five sites, a major lateral haplotype (n = 7) representing four sites and five single-sample haplotypes (n = 1 each). In U. ohnoi, the center (n = 42), representing Shinyang and Pyoseon, connected to two haplotypes (n = 1 each) as well as one (n = 1) from Shinyang (Fig. 6F). The haplotype networks of rbcL in the novel species, U. australis, and U. lactuca showed fewer variations than those of tufA (Supplementary Fig. S1). In the haplotype networks of rbcL, U. austrlais and U. ohnoi showed the greatest variations. U. australis was composed of five haplotypes containing the center (n = 26) from five sites, a major lateral haplotype (n = 9) from three sites and three single-samples (n = 1 each) from two sites (Supplementary Fig. S1). On the other hand, U. ohnoi was composed of the center (n = 41) from two sites and four single-samples from two sites (n = 1 from Pyoseon; n = 1 from Shinyang each) (Supplementary Fig. S1). U. lactuca and the novel species exhibited two haplotypes composed of the center (n = 24 of U. lactuca; n = 33 of the novel species) and each single-sample haplotype (Supplementary Fig. S1). The haplotypes of U. laetevirens were three consisting of the center (n = 25) from five sites and two single-samples from two sites (n = 1 each) (Supplementary Fig. S1).
Ulva pseudo-ohnoi H. W. Lee, J. C. Kang & M. S. Kim sp. nov. (Fig. 3A, B & D–F)
Holotype
MSK-U41-JD-SM-D-03 (Fig. 3A), vegetative, Jongdal, Jeju Island, Korea, Aug 21, 2017, deposited in the herbarium of JNUB.
Isotype
MSK-U42-JD-SM-D-04, MSK-U43-JD-SM-D-05, MSK-U44-JD-SM-D-06, MSK-U45-JD-SM-D-07, MSK-U50-JD-SM-L-12, MSK-U51-JD-SM-L-13, Jongdal, Jeju Island, Korea, Aug 21, 2017 (deposited in JNUB).
Etymology
The specific epithet, pseudo-ohnoi, was selected due to morphological and habitat similarity with U. ohnoi.
Habit and vegetative morphology
U. pseudo-ohnoi H. W. Lee, J. C. Kang & M. S. Kim sp. nov. is similar in habit to U. ohnoi, as a foliose thallus with ruffled margins and extremely irregular expansion during growth (Fig. 3A & B). Thallus grows 25–30 cm high, up to a meter. Thickness of thallus is 50–80 μm, up to 90 μm. Its thallus is delicate in texture and easily torn, and often shows splits in the upper portion, like U. ohnoi (Fig. 3A & B). Its color ranges from light pale green to moderately dark green (Fig. 3A & B). Its fronds may have a few small irregular perforations (Fig. 3B). Its margins have short and multifid teeth with bluntly rounded to concave apexes (Fig. 3D). Cells in surface view are cuboidal to roundish (Fig. 3E). Its thallus is distromatic (Fig. 3F), and cell profiles in a cross-section are cuboidal (Fig. 3F). In the present study, U. pseudo-ohnoi sp. nov. is difficult to distinguish from U. ohnoi using only habit characteristics and morphological observations. Most individuals of U. pseudo-ohnoi sp. nov. are free-floating at bloom sites like U. ohnoi.
Remarks
This novel species was first recognized as a cryptic species from Japan, characterized by microscopic marginal teeth similar to those of U. conglobata, and with a close phylogenetic relationship to U. laetevirens based on rbcL and internal transcribed spacer (ITS) regions (Matsumoto and Shimada 2015). In addition, this organism was reported from the coast of Jeju Island, Korea (Kang et al. 2019). U. pseudo-ohnoi sp. nov. is difficult to differentiate from U. ohnoi morphologically due to similar characteristics including a greatly expanded thallus, microscopic marginal teeth, and free-floating habit (Hiraoka et al. 2003) (Fig. 3). However, our concatenated ML analysis of the tufA and rbcL dataset indicates that U. pseudo-ohnoi sp. nov. is paraphyletic to U. ohnoi (Fig. 2), and this finding is supported by previous ITS phylogenetic analyses (Matsumoto and Shimada 2015, Kang et al. 2019).
Ulva lactuca Linnaeus 1753, p. 1163 (Fig. 4A–C & E–G)
Distribution
Korea (Bae 2010, Kim et al. 2013); Japan, Australia, Hawaii, Indo-Pacific region, The Mediterranean Sea, Central Atlantic (Hughey et al. 2019).
Habit and vegetative morphology
U. lactuca Linnaeus has various habit characteristics, including a lobed thallus with ruffled margins (Fig. 4A), highly lobed thallus around a holdfast with several perforations (Fig. 4B), or an irregularly furcated thallus with a number of long and narrow lobes (Fig. 4C). Thallus grows 10–15 cm high, up to 30 cm, and 45–90 μm thick, up to 100 μm. Its blade is medium green when young (Fig. 4A) to olive green when old (Fig. 4B & C). Its margins have protuberant, simple, mild and very short teeth, sometimes multifid, with blunt apices (Fig. 4E). Cells in surface view are cuboidal to polygonal (Fig. 4F). Its thallus is distromatic (Fig. 4G). Cell profiles in a cross-section include bluntly rounded cells with some acutely pointed apices (Fig. 4G). In the present study, most individuals of U. lactuca were attached to rocks at bloom sites.
Remarks
U. lactuca was described on the basis of the epitype of U. fasciata (UC2050475) that supports the lectotype (PC0119343) from New Port, Alexandria, Egypt by Delile including the holotype of U. fenestrata from Kamchatka, Russia by Postels and Ruprech (Hughey et al. 2019). Although it is one of the most cosmopolitan species found in all temperature conditions (Hughey et al. 2019), the true taxonomic position of U. lactuca was only recently revealed through advanced analysis of the lectotype specimen for variations of tufA and rbcL. That analysis indicated that the type specimen of U. fasciata Delile first reported from Alexandria, Egypt and specimens recognized as U. fasciata collected worldwide are all U. lactuca, while numerous specimens recognized to date as U. lactuca should be considered U. fenestrata Postels & Ruprecht (Hughey et al. 2019). In the Ulva bloom areas of Jeju, this species is always the predominant species in conjunction with U. ohnoi, U. australis, U. laetevirens, and U. pseudo-ohnoi sp. nov. We observed that U. lactuca grows in the upper intertidal and mid-tidal zones mainly as an attached plant but also in a free-floating form, and exhibits maximum growth during late summer.
Ulva laetevirens Areschoug 1854, p. 370 (Fig. 5A–C & E–G)
Distribution
Korea (this study); North America (Kirkendale et al. 2013); Australia and New Zealand (Kraft et al. 2010, Kirkendale et al. 2013); Africa (Miladi et al. 2018); Europe (Miladi et al. 2018).
Habit and vegetative morphology
U. laetevirens is irregularly contoured and lobed with small perforations (Fig. 5A–C). Its thallus is medium green to green (Fig. 5B & C), and sometimes fairly dark green (Fig. 5A). Its fronds are broadly palmate to irregularly lobed (Fig. 5A & B) or irregularly furcated and expanded (Fig. 5C). Thallus ranged from 15–30 cm high. Thallus thickness is 70–85 μm. Its fronds have multifid, sharp, long and irregularly protuberant marginal teeth (Fig. 5E). Marginal denticulation is simple to multiple ordered by growing (Fig. 5E). Cells in surface view are unordered or show both longitudinal and transverse alignments, and have cuboidal or sub-rectilinear shapes with roundish outlines (Fig. 5F). Its thallus is distromatic (Fig. 5G). In cross-section, most cells are bluntly rounded, with some sharply tapered to acutely pointed apices (Fig. 5G). In the present study, most individuals of U. laetevirens were attached to rocks at bloom sites.
Remarks
An U. laetevirens specimen was collected from Port Phillip, Victoria, Australia (Areschoug 1854). This species was also recently reported in Long Island Sound, along the northwest Atlantic Ocean in North America (Mao et al. 2014), as well as in several Mediterranean coastal countries since the late 1990s (Sfriso 2010). U. laetevirens exhibits a conical cell shape in the rhizoidal and basal regions, without marginal teeth along the basal and median regions (Sfriso 2010). Mao et al. (2014) partially sequenced the plastid elongation factor gene (tufA) and placed specimens from Holly Pond, USA in a well-supported clade with published sequences of U. laetevirens (HQ610428) identified previously with no divergence. Kang et al. (2019) described U. laetevirens inhabiting Jeju Island using tufA and ITS analyses. Based on our tufA and rbcL DNA barcoding methods, the presence of U. laetevirens around Jeju Island was further confirmed (Fig. 2). This species showed the most strongly developed microscopic marginal teeth (Fig. 5) compared to U. lactuca, U. ohnoi, and U. pseudo-ohnoi sp. nov.
Ulva ohnoi M. Hiraoka & S. Shimada 2003, p. 20, Figs 1–22 (Fig. 6D & E)
Distribution
Korea (Bae 2010, Kim et al. 2013); Japan (Hiraoka et al. 2003); America (Melton et al. 2016, Chávez-Sánchez et al. 2018); Southwest Asia (Kazi et al. 2016, Krupnik et al. 2018); Australia and New Zealand (Kirkendale et al. 2013); Pacific Islands (O’Kelly et al. 2010); Africa (Miladi et al. 2018); Europe (Miladi et al. 2018).
Habit and vegetative morphology
Ulva ohnoi M. Hiraoka & S. Shimada is distromatic. Its fronds are foliose and ruffled (Fig. 6D & E). Its thallus is light green or yellowish green in color (Fig. 6D & E). Its blade is easily torn and fragile, and is often split in the upper portion. Some individuals are lightly and irregularly perforated (Fig. 6E). Its fronds generally grow up 40 cm in length and 20 cm in width (Fig. 6D & E), and may expand to over 1 m long by 1 m wide. In the present study, most individuals of U. ohnoi were free floating at bloom sites.
Remarks
U. ohnoi Hiraoka et Shimada was described from southern and western Japan, and is characterized by large, fragile, easily-torn thalli with microscopic marginal teeth and 1–3 pyrenoids (Hiraoka et al. 2003). This species is known to form green tides (Kawai et al. 2007, O’Kelly et al. 2010, Kirkendale et al. 2013, Melton et al. 2016) and has been introduced to Italy, Spain, Australia, North and South America, India, and the Hawaiian Islands (Miladi et al. 2018). In Japan, the green tides are caused by free-floating thalli of U. ohnoi, and huge biomass of this species occurs in some calm bays (Hiraoka et al. 2003). We found that U. ohnoi is the dominant species along the eastern coast of Jeju Island, such as Shinyang, which experiences a massive bloom every year, and Pyoseon, which has potential for bloom development (Fig. 6F). Our specimens showed microscopic marginal teeth. According to the results of Hiraoka et al. (2003), the presence or absence of marginal teeth reflect the phylogeny of Ulva, and this feature provides a useful genetic morphological characteristic.
Ulva australis Areschoug 1854, p. 370 (Fig. 6A & B)
Habit and vegetative morphology
Ulva australis Areschoug has distromatic blades. Generally, its fronds are foliose, lobed, and slightly ruffled (Fig. 6A & B), but may be robust, more ruffled, and slightly dentate (Fig. 6B). Its fronds are irregularly perforated (Fig. 6A & B). Its thallus color is yellowish green to grass green (Fig. 6A & B). Its thallus size is normally 10–15 cm long by 10–15 cm wide (Fig. 6A & B), but can grow up to 20 cm in length or width and its shape varies among individuals (Fig. 6A & B). The blades are attached with a single basal holdfast or free floating. Free-floating individuals are more expanded than attached ones. In the present study, most individuals of U. australis were attached on various substrates at bloom sites.
Remarks
Ulva australis was originally reported as a specimen from Port Adelaide, South Australia and is conspecific with the younger name U. pertusa Kjellman (Kraft et al. 2010). These two species are consistent, with characteristics including thallus thickness up to 300 μm near the base, 1–2 pyrenoids per cell, and fronds that are irregularly divided from the base (Couceiro et al. 2011). Several Ulva species, including U. australis in Australia, showed little genetic variation across large geographic distances, indicating they are cosmopolitan species (Kirkendale et al. 2013). Similarly, in the present study, intraspecific variations within U. australis among our specimens, including Australian and cosmopolitan ones, were small, with ranges of 0–0.4% in tufA and 0–0.2% in rbcL. This species showed the widest distribution and greatest number of haplotypes among the five bloom-forming species analyzed (Fig. 6C).
DISCUSSION
We provide molecular-assisted taxonomic results for Ulva species collected from various coastal environments of Jeju Island, including at Shinyang, an embayment site that experiences severe massive blooms annually (Song et al. 2016), and Jocheon, a harbor affected by artificial structures (Kwon et al. 2017). The species composition at massive Ulva bloom sites around Jeju Island comprises: (1) U. australis Areschoug, (2) U. lactuca Linnaeus, (3) U. laetevirens Areschoug, (4) U. ohnoi M. Hiraoka & S. Shimada, and (5) U. pseudo-ohnoi H. W. Lee, J. C. Kang & M. S. Kim sp. nov. (Fig. 2). These species are characterized by distromatic and foliose thalli with attached and / or free-floating habits. Excluding U. australis, four of these species formed a monophyletic clade sharing unique morphological characteristics, such as marginal dentation (Figs 3–5) (Hiraoka et al. 2003). Although this monophyletic group exhibited low interspecific divergence, 0.6–2.5% for tufA and 0.4–0.8% for rbcL, each specific clade could be clearly distinguished with both DNA markers. The tufA gene proposed for DNA barcoding of green macroalgae (Saunders and Kucera 2010) provides a powerful tool for delimiting species boundaries due to its high interspecific divergence (Lee and Kim 2015). In the present study, the interspecific divergence of the tufA marker in the genus Ulva was higher than that of rbcL, and was sufficient to distinguish among the four species of Ulva with similar morphological traits.
For molecular study of Ulva, the nuclear ITS and the plastid rbcL regions have been employed to reveal phylogenetic relationships and to delimit species diversity (Malta et al. 1999, Shimada et al. 2003, Hayden and Waaland 2004, Loughnane et al. 2008, Kraft et al. 2010, O’Kelly et al. 2010). Because of poor amplification for the ITS region (Saunders and Kucera 2010) and low levels of genetic diversity for rbcL region hampering species-level resolution (Heesch et al. 2009), the tufA of chloroplast was proposed as a more successful marker having species-level resolution than ITS and rbcL (Saunders and Kucera 2010). Molecular data from tufA marker showed more interspecific divergence for the genus Codium (Lee and Kim 2015). In the present study, the plastid marker tufA also provided high resolution for species boundaries between Ulva species having marginal denticulation, U. lactuca, U. laetevirens, U. ohnoi, and U. pseudo-ohnoi sp. nov. with higher interspecific divergence than rbcL (Supplementary Fig. S2).
Although Kang et al. (2019) conducted the molecular study of Ulva along the coast of Jeju Island, Korea based on the nuclear ITS and the plastid tufA regions, it was hard to resolve the species delimitation between U. ohnoi and the closely related species having marginal denticulation. By reassessing the species boundaries of them on tufA region including the sequences obtained in the present study (Supplementary Table S3), the four genetic groups were distinguished distinctly, the true U. ohnoi clade, the U. pseudo-ohnoi clade and an unidentified clade (Supplementary Fig. S2). Extended field samplings would clarify the species entities and delineate species diversity so far only encountered in single samples previously such as U. pseudo-ohnoi sp. nov. (Heesch et al. 2009, Kirkendale et al. 2013). The increasing tufA application for species delimitation can derive the deeper understanding of the species diversity of the genus Ulva through additional sampling.
Matsumoto and Shimada (2015) first recognized our new species, U. pseudo-ohnoi sp. nov., as Ulva sp. 1 based on the ITS and rbcL analyses. They described that this species seemed most similar to U. conglobata Kjellman morphologically (Matsumoto and Shimada 2015). However, they did not provide the molecular proofs of U. conglobata. In order to assign Ulva sp. 1 (= U. pseudo-ohnoi sp. nov.) as U. conglobata, it is necessary to analyze the specimens of U. conglobata by molecular approach preferentially because of morphological confusion to delimit species boundaries. U. pseudo-ohnoi was closely related to U. spinulosa and U. reticulata Forsskål (Matsumoto and Shimada 2015). By Monotilla et al. (2018), U. reticulata was already not conspecific with U. spinulosa and U. ohnoi based on ITS analysis. In the U. reticulata clade of Monotilla et al. (2018), the specimens of U. reticulata was collected from Philippiness (Monitilla et al. 2018). Monotilla et al. (2018) did not analyze the type materials from the type locality where is Saudi Arabia and Yemen (Silva et al. 1996). Therefore, it is hard to identify these specimens as the true U. reticulata taxonomically. Therefore, the specimens of U. reticulata from Philippiness would be assigned to U. pseudo-ohnoi sp. nov.
Historically the identification of Ulva species in Korea has been conducted almost entirely based on morphology, and only a few studies have employed molecular analysis of this genus (Bae and Lee 2001, Kang et al. 2014, 2019). Kang et al. (2019) reported the proportional occurrences of Ulva species using molecular approaches. We determined that the most problematic Ulva bloom, which occurs in Shinyang, is composed of four distromatic and foliose species, U. australis, U. lactuca, U. laetevirens, and U. ohnoi (Table 1), that have attached and free-floating habits. Another massive bloom in Jocheon is caused by U. australis, U. laetevirens, and U. pseudo-ohnoi (Table 1). The species composition of Ulva blooms at the embayment sites, Shinyang and Pyoseon, was distinguished in the harbor sites, Jecheon and Jongdal, distinctly. U. australis, U. lactuca, U. laetevirens, and U. ohnoi were causative for blooms at the embayment, but blooms at harbor were composed of U. australis, U. laetevirens, and U. pseudo-ohnoi sp. nov. in the present study. Moreover, possible bloom areas affected by coastal features or artificial modifications have high risk of massive invasions by one or more of the five Ulva species: U. australis, U. lactuca, U. laetevirens, U. ohnoi, and U. pseudo-ohnoi sp. nov.
In our study around Jeju Island, the most common species in the Ulva bloom area during all seasons were U. ohnoi, U. australis, and U. pseudo-ohnoi sp. nov., followed by U. laetevirens and U. lactuca (Table 1). Although U. australis (as U. pertusa) has been reported from southern Asia (Silva et al. 1996), these results would likely be identified as U. ohnoi based on molecular data, as U. australis is rarely distributed in subtropical regions such as the Ryukyu Islands (Hiraoka et al. 2003, Hanyuda et al. 2016). U. australis has seven haplotypes for tufA and five for rbcL, showing more variation in Northeast Asia, Korea, and Japan than in other areas (Hanyuda et al. 2016). In Japan, U. ohnoi causes green tides in warm temperature regions along the southern and western coasts (Hiraoka et al. 2003). Therefore, massive blooms of free-floating Ulva in high-temperature regions can be assumed to be U. ohnoi introduced from subtropical habitats (Hiraoka et al. 2003).
Along the eastern coast of Jeju Island, all blooms of Ulva were previously identified as U. conglobata Kjellman and U. pertusa Kjellman, which is a synonym of U. australis (Kwon et al. 2017). However, we revealed that the dominant massive free-floating species are U. ohnoi. Although U. australis is also a bloom-forming species, it is not a free-floating, but requires attachment around Jeju Island. On the other hand, green tides in the Yellow Sea of the Korean Peninsula consisted predominantly of U. linza and rarely of U. compressa or U. prolifera (Kang et al. 2014). U. linza is characterized by strap-shaped, sheet-like thalli with ruffled blade margins, but it shows considerable phenotypic plasticity induced by the environment (Brodie et al. 2007, Belton et al. 2014). Most floating bloom species from the western coast of Korea were confirmed to be U. linza based on their rbcL sequences, but manifested U. prolifera-type morphology (Kang et al. 2014). In the present study, the molecular phylogenetic tree showed different main species among free-floating Ulva blooms, including U. ohnoi with U. pseudo-ohnoi sp. nov. around Jeju, whereas U. compressa, U. flexuosa and LPP compex (U. linza-U. procera-U. prolifera) along the west coast of Korea (Duan et al. 2012, Zhao et al. 2013).
Ulva blooms are becoming a regular and increasingly severe problem in estuarine systems and coastal areas worldwide. Therefore, the present assessment and characterization of Ulva blooms will provide information to decision-makers to guide the management of green macroalgal blooms while considering the Ulva seaweed industry as a potentially emerging market.
SUPPLEMENTARY MATERIALS
Supplementary Table S1.
Analyzed specimens of Ulva species causing blooms around Jeju Island in this study (https://www.e-algae.org).
Supplementary Table S2.
The representative sequence datasets for combined maximum likelihood analysis of tufA and rbcL of Ulva from GenBank including outgroups (https://www.e-algae.org).
Supplementary Table S3.
The tufA identification of Ulva species having marginal denticulation from GenBank based on DNA barcoding (https://www.e-algae.org).
Supplementary Fig. S1.
Statistical parsimony haplotype networks of rbcL marker generated in TCS within each Ulva blooming species from Jeju Island, Korea. Each haplotype is represented by a circle, and the size of each circle is proportional to the number of individuals with that haplotype (https://www.e-algae.org).
Supplementary Fig. S2.
Neighbor-joining analysis of Ulva species having marginal denticulation based on the plastid tufA. Branch lengths are proportional to substitution rate (https://www.e-algae.org).