INTRODUCTION
Aquaculture of marine algae is an important industry, especially in Asia. The production of seaweeds more than doubled between 2000 and 2012 (Food and Agriculture Organization of the United Nations 2014). The red alga Pyropia is the most consumed alga in the world, both for food and in the biomedical industry (e.g., porphyran, pycobilliproteins) (Gachon et al. 2010). In 2013, Pyropia made up about 1.8 million tons which is about 8% of the total global seaweed production, with values of US $1.2 billion (FAO FishStat et al. 2016).
Pyropia cultivation losses amount to over US $10 million annually from different diseases (Gachon et al. 2010, Blouin et al. 2011, Kim et al. 2014). Diseases like green-spot disease and Olpidiopsis blight as well as red-rot disease, result in a great decrease in productivity, yield and crop value (Kawamura et al. 2005, Klochkova et al. 2012, 2016b, Kim et al. 2014, 2016). With increasing farming intensity and increasing temperatures, caused by global warming, disease severity and occurrence is also expected to increase (Ding and Ma 2005, Gachon et al. 2010).
The most common diseases of cultivated Pyropia are the red-rot disease and the “Olpidiopsis blight,” caused by the oomycetes Pythium porphyrae M. Takah. & M. Sasaki and Olpidiopsis pyropiae G. H. Kim & T. A. Klochkova, respectively (Kim et al. 2014, Klochkova et al. 2016b). Algal pathogens of Pyropia have been reported for a closely related species, Pythium chondricola De Cock (Lee et al. 2015), an oomycete first described from Chondrus crispus Stackhouse in the Netherlands (De Cock 1986). These organisms are of particular concern as they are the main cause of harvest loss in China, Korea, and Japan (Kim et al. 2014). Red-rot disease alone leads to a yield reductions of up to 20% in farms in the Ariake Sea, Japan (Kawamura et al. unpublished data) and causes death of the host within a few days (Ding and Ma 2005).
Oomycota are fungal-like eukaryotes that are classified in the Stramenopiles (Patterson 1989). The genus Pythium is found throughout the world in freshwater, marine, and terrestrial habitats (Kageyama 2014, Klochkova et al. 2016a). Species delimitation is difficult in these microorganisms based solely on morphology, while accurate species delimitation is critical for nearly all biological applications. For example, incorrect names may lead to misguided efforts to eradicate two species while only one exists. This has led to the increased use of molecular data to identify and delimit species (e.g., Sandoval-Sierra et al. 2014, De la Bastide et al. 2015). Several methods have been developed to delimitate species using molecular data and to discover putative species, and a multi-pronged approach has been recommended as all methods are based on different assumptions (Leliaert et al. 2014). The application of names is further complicated by culture collections and DNA databases containing misassigned sequences (Sandoval-Sierra et al. 2014)
Pythium was first recorded on Porphyra tenera (now Pyropia tenera) in Japan (Arasaki 1947), but it still took three decades to designate the infective agent as Pythium porphyrae (Takahashi et al. 1977). Recent molecular studies, using mitochondria-encoded cytochrome oxidase subunit 1 (COI) and nuclear-encoded internal transcribed spacer of the rDNA cistron (ITS), have shown that Pythium consists of several well-supported clades with the marine species P. porphyrae and P. chondricola being closely related and in the same clade (clade A) (Lévesque and De Cock 2004, Robideau et al. 2011).
New Zealand offers diverse habitats for a range of Pyropia and other closely related Bangiales, with more than 30 species found there (Nelson et al. 2006, Sutherland et al. 2011). Little is known about Pythium infection in Pyropia in natural habitats, even though the organism is well investigated in aquacultural settings (e.g., Park et al. 2000, Kawamura et al. 2005, Kim et al. 2014). A few studies have explored the host-range of Pythium in marine algae (e.g., Klochkova et al. 2016a) and generally indicate that they can have wide host ranges. Due to the wide variety of host substrate-specific relationships and their huge economic impact, the clarification of host-parasite relationships and proper identification of these pathogens is warranted.
We isolated a Pythium species infecting Pyropia plicata W. A. Nelson in Wellington, New Zealand and performed susceptibility tests using diverse algae, especially species of Bangiales. Species delimitation analysis and morphological investigation showed that synonimization between Pythium porphyrae and P. chondricola is necessary.
MATERIALS AND METHODS
Sampling and isolation
All samples of Pyropia plicata were collected at Moa Point Wellington, New Zealand (41°20′31.6″ S, 174°48′35.4″ E) on three different days: Jun 2, Jun 9, and Jul 5, 2016. The blades were checked for infection under the dissecting microscope. Sections that were infected were cut out (approximately 5 × 5 mm) and placed in wells with sterilized Provasoli’s Enriched Seawater (1/4 strength PES, 36 PSU) (West and McBride 1999), to which a few drops of GeO2 (25 μg mL−1) and a few mg of Penicillin were added. The 6-well-boxes with the samples were maintained in an incubator at 15°C and 12 : 12 light : dark cycle (20 μmol m−2 s−1). Once a week the medium was changed. Every three to five days uninfected Pyropia plicata tissue, which was collected from the same location, was added to maintain the cultures.
To isolate the oomycete from the algal tissue, the tissue was briefly wiped with either ethanol (70%) or commercial bleach (0.5%) to clean the surface of potential contaminants. The tissue was then placed in either flasks containing 100% cornmeal-seawater medium or on 50% and 100% cornmeal-seawater agar plates. The seawater-cornmeal medium was prepared as follows: 50 g of organic cornmeal was added to an equal amount of sterile seawater (36 psu) and heated to boiling. After simmering for 10 min, the solution was cooled and allowed to settle, it was filtered through cheese cloth and autoclaved. Before adding oomycetes, 200 μL of streptomycin (10 mg mL−1 ethanol) and 200 μL of rifampin (4.5 mg mL−1 methanol) were added to every 50 mL of medium. Agar plates were made with 50% and 100% seawater with 17 g L−1 BBL Corn Meal Agar (BD Biosciences, Auckland, New Zealand), autoclaved and poured into plastic petri dishes. Before inoculation the plates were spread with 100 μL of streptomycin (10 mg mL−1 ethanol) and 100 μL of rifampin (4.5 mg mL−1 methanol) to prevent bacterial growth (Francis et al. 2016). The plates and flasks, after inoculation, were kept in an incubator at 15°C.
To induce reproduction in Pythium, we prepared Arasaki B Medium (Arasaki et al. 1968) and added the isolated hyphae from the agar plates or cornmeal media to the solution. These cultures were incubated at 20°C.
Host specificity
For infection and host specificity experiments, different algae were collected at Moa Point, Wellington. Newly collected healthy tissue of Pyropia plicata (the original host) and various other algae (Table 1) were added individually to Pyropia plicata infected with Pythium and placed together in wells with sterilized 1/4 strength PES, with added GeO2 (25 μg mL−1) and Penicillin, and incubated at 15°C and 12 : 12 light : dark cycle (20 μmol m−2 s−1) for up to 2 weeks. Cultured conchocelis stages were also tested for infectivity (from Wendy Nelson, National Institute of Water and Atmospheric Research, Wellington, New Zealand) (Supplementary Table S1). Tissue was monitored microscopically every two to three days. All microscopic observations were made with a Olympus microscope (BX63 F; Olympus, Tokyo, Japan) and attached DB80 Camera.
Molecular identification
The Bangiales were extracted using the Chelex extraction method (Goff and Moon 1993). The oomycetes were extracted using a modified cetyltrimethylammonium bromide (CTAB) extraction method (Zuccarello and Lokhorst 2005). DNA was stored at −20°C.
To molecularly identify the Bangiales, we used the large subunit ribulose-bisphosphate carboxylase / oxygenase (rbcL) gene. RbcL was amplified in two parts: part 1 using primers F145 (Kim et al. 2010) and R753 (Freshwater and Rueness 1994); and part 2 using primers F753 and R-rbcS-start (Freshwater and Rueness 1994). For the amplification of Pythium, two different primer pairs were used: the oomycete specific COI primers (forward OomCox1-Levup, reverse OomCox1-Levlo), and occasionally an alternate reverse primer, Fm85mod (Robideau et al. 2011) and for the ITS the universal primers, ITS1 and ITS4 (White et al. 1990).
The polymerase chain reaction (PCR) reaction volume was 30 μL, containing 1× reaction buffer (Bioline Reagents Ltd., London, UK), 0.2 μM dNTP’s, 2.5 mM MgCl2, 0.033% BSA, each 0.25 pmol of forward and reverse primer, 1 U BIOTaq DNA Polymerase (Bioline Reagents Ltd.) and 1 μL of template DNA. The PCR program (PTC-100; MJ Research Inc., Watertown, MA, USA) was 5 min at 95°C, followed by 1 min at 94°C, 1 min annealing, 1 min at 72°C for 36 cycles and final extension step for 5 min at 72°C. The annealing temperature for the rbcL primers was 45°C, for the COI primers 53°C and for the ITS primers 50°C. The PCR products were electrophoresed in 1.0% agarose gels and visualized using ethidium bromide. Samples of appropriate size and intensity were prepared for sequencing using ExoSAP-IT following standard protocols (USB product; Affymetrix, Santa Clara, CA, USA). Sequencing was done commercially (Macrogen Inc., Seoul, Korea).
Phylogenetics
Forward and reverse sequences were assembled, edited and consensus sequences were generated in Geneious 9.1 (http://www.geneious.com) (Kearse et al. 2012). To confidently identify the Bangiales collected, newly generated large subunit ribulose-bisphosphate carboxylase / oxygenase (rbcL) sequences were added to a data set of all Bangiales (Sutherland et al. 2011) for phylogenetic confirmation of species identification. Smithora naiadum (C. L. Anderson) Hollenberg, Chlidophyllon kaspar (W. A. Nelson and N. M. Adams) W. A. Nelson and Pyrophyllon subtumens (J. Agardh ex R. M. Laing) W. A. Nelson, as members of the Erythropeltidales, were used as outgroups as in Sutherland et al. (2011).
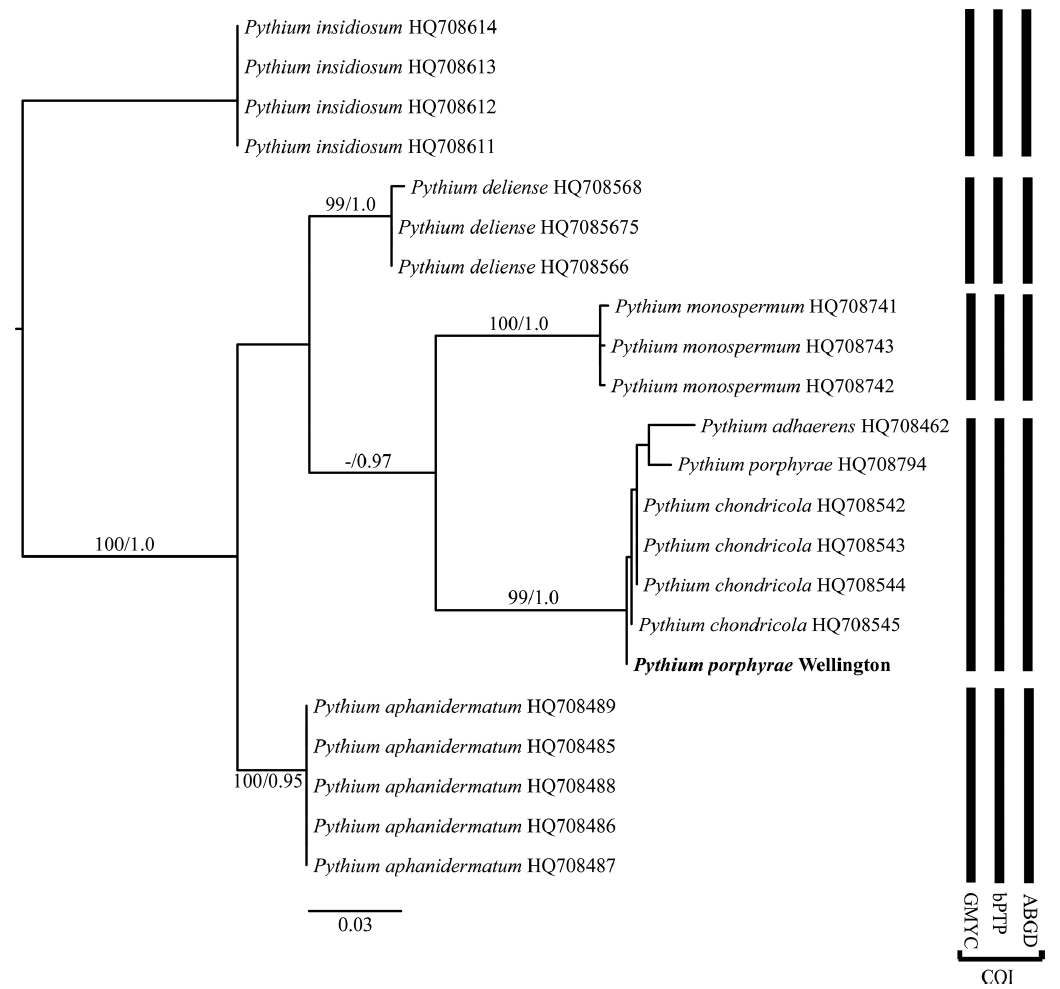
Data sets were produced (for both COI and ITS) for our isolated Pythium and sequences downloaded from GenBank of “clade A” species of Pythium, a supported subclade within Pythium containing red alga pathogens (Robideau et al. 2011). P. insidiosum De Cock, Mendoza, Padhye, Ajello & Kaufman, a member of Pythium clade C (Robideau et al. 2011), was used as an outgroup (Supplementary Table S2).
The phylogenetic tree for the Bangiales was constructed with maximum-likelihood (ML) using RAxML 7.2.8 (Stamatakis 2006) under the GTR + gamma model with partitioned codons. Support for individual nodes was determined by 400 bootstrap replicates. Phylogenetic trees for Pythium were constructed with ML and Bayesian inference (BI). ML analyses were performed using RAxML 7.2.8 under the GTR + gamma model, with codons partitioned for the COI data. Support for individual nodes was determined by 1,000 bootstrap replicates. BI analyses were conducted using MrBayes 3.2 (Ronquist et al. 2012) under the GTR + gamma model for 3 million generations with two independent runs, a sampling frequency of 1,000 and a burn-in of 300 trees.
Species delimitation
To delimit species in clade A, especially the species Pythium chondricola and P. porphyrae, we used three methods: distance-based (Automatic Barcode Gap Discovery [ABGD], web last modified 17/09/2016) (Puillandre et al. 2012), a tree-based method (Poisson-Tree processes [PTP]) (Zhang et al. 2013) and a model-based method (Generalized Mixed Yule Coalescent [GMYC]) (Pons et al. 2006, Monaghan et al. 2009, Fujisawa and Barraclough 2013). All analyses were run with the COI and ITS datasets. ABGD was run with the following settings: Pmin = 0.001, Pmax = 0.1, steps = 10, X (relative gap width) = 1.0, Nb bin (distance distribution) = 20 and Jukes-Cantor (JC69) or Kimura (K80) parameter models. We used the Bayesian variant of the PTP method (bPTP). The current version of the software (Oct 2016) used our RAxML COI and ITS gene tree with 100,000 Markov chain Monte Carlo (MCMC) generations and thinning every 100. The ultrametric tree for GMYC was generated with BEAST 1.8.2 (Drummond et al. 2012) from COI and ITS alignments after removing identical sequences. A coalescent constant size tree prior (Kingman 1982) was set under an uncorrelated lognormal relaxed clock and GTR + gamma + invariant sites model. The analysis was set up for 50 million generations and a sampling frequency of 5,000. Before performing the GMYC analyses, we checked the estimated samples size with Tracer 1.6 (Rambaut et al. 2014). The maximum clade credibility tree was computed using TreeAnnotator 1.8.3 (Drummond et al. 2012). The resulting ultrametric tree was imported into the GMYC web server, running the single threshold (sGMYC) (Pons et al. 2006).
RESULTS
Molecular identification
All tested species of Bangiales were identified by BLAST searches and phylogenetic analysis of rbcL, and were 100% identical to sequences already present in New Zealand and known from the Wellington area (Sutherland et al. 2011) (Supplementary Fig. S1). The only host of Pythium porphyrae in Wellington was Pyropia plicata.
Samples of infected Pyropia were detected at Moa Point, especially in the austral winter month, Jun–Aug (Fig. 1). All collected Pythium samples from Moa Point (Supplementary Table S3) had identical COI (Genbank accession No. KY650705) and ITS (Genbank accession No. KY630550) sequences (i.e., sequences from blades and all Pythium cultures).
The phylogenetic analyses, both ML and BI, using ITS and COI showed that our samples formed a well-supported clade with Pythium chondricola, P. porphyrae, and P. adhaerens. In the COI analysis (Fig. 2) the support for this clade was high and this clade was a moderately supported sister clade (0.97 Bayesian PP) to P. monospermum. The level of variation within the P. adhaerens–P. chondricola–P. porphyrae clade was low. The ITS analysis was very similar (Fig. 3) and again showed a well-supported clade of P. adhaerens–P. chondricola–P. porphyrae. There was slight variation in P. adhaerens from the identical sequences of P. chondricola and P. porphyrae.
All three species delimitation methods, using the COI datasets, indicated five species, and placed P. chondricola, P. porphyrae, and P. adhaerens within the same putative genetic species (Fig. 2). The species delimitation results of the ITS datasets were very similar but both the PTP and AGBD methods separated P. adhaerens from P. chondricola and P. porphyrae (Fig. 3).
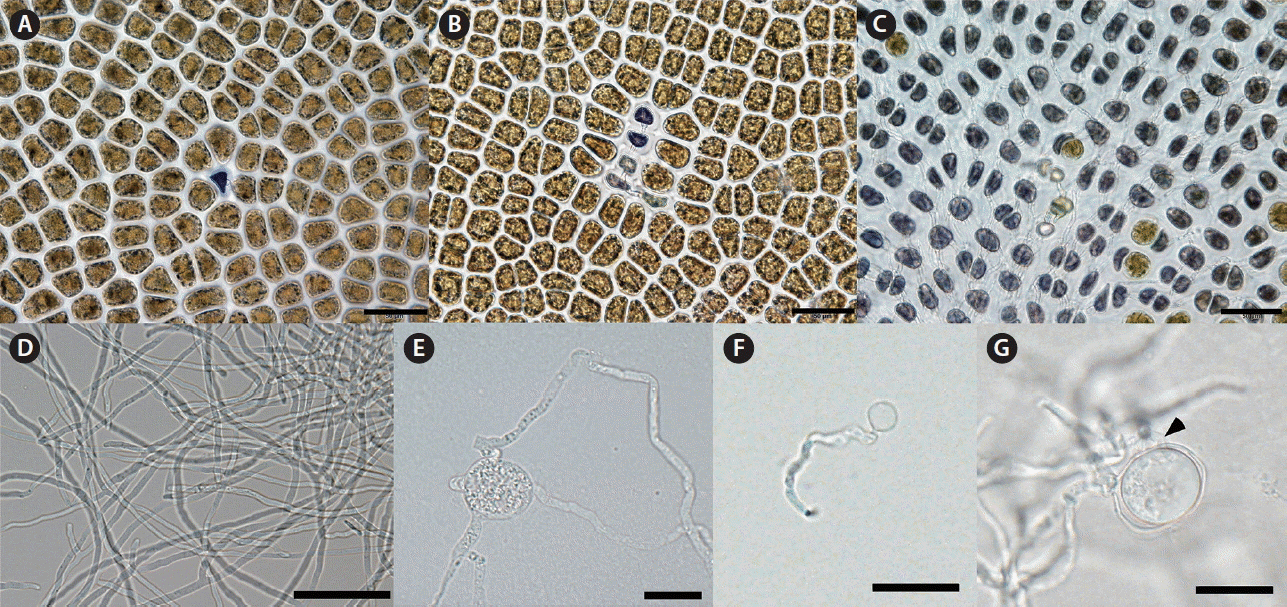
The infection process of P. porphyrae on its host Pyropia plicata led to the dark coloration of infected cells and the quick spread of the infection, by hyphae, throughout the tissue (Fig. 4A–C). Pythium porphyrae isolated into culture media, produced hyphae between 2.5–5 μm wide (Fig. 4D). On agar plates hyphae produced hyphal swellings (Fig. 4E). Although zoosporangia were not observed in our cultures we did see spores in the culture media that had germinated (Fig. 4F). Sexual reproduction was only observed a few times in Arasaki medium, oosporangia were smooth, occasionally slightly undulating, with one plerotic smooth oospore with a single antheridium (Fig. 4G).
Host specificity
Several, both gametophytes and sporophytic ‘conchocelis-stages,’ of Bangiales, plus other algae were tested for infection with P. porphyrae (Table 1). The original host of P. porphyrae in Wellington, Pyropia plicata, was newly infected within 24 h. After two weeks, an approximately 5 × 5 mm piece of tissue was dead in our culture conditions. Other Pyropia and Porphyra species could be infected within less than 48 h (Fig. 5A–C). The infection process, while appearing slightly slower than that from the original host, was morphologically fairly similar (Fig. 5). Even after more than one week the gametophytic thalli of ‘Bangia’ 2 sp. BGA, ‘Bangia’ 1 sp. BMW, Champia novae-zealandiae, Bostrychia arbuscula, and Ulva sp., showed no infection with P. porphyrae (Table 1). We did not get any observable infection in any conchocelis phases.
DISCUSSION
All Pythium found and isolated in Wellington had identical COI and ITS sequences. These sequences were identical to the type culture of P. chondricola, isolated from Chondrus crispus in the Netherlands and very similar to sequences of P. porphyrae. Our species delimitation methods indicate that P. chondricola, P. porphyrae, and P. adhaerens are either the same species (COI analysis), or P. adhaerens is possibly a different species (ITS analysis). Also morphologically, no substantial difference can be seen between descriptions of P. chondricola, P. porphyrae, and our isolate from New Zealand (Table 2). Correct species identification is critical for all further biological analysis. For example, if two pathogens are recognized on cultivated Pyropia (Pythium porphyrae and P. chondricola) (e.g., Kim et al. 2014, Lee et al. 2015), this could interfere with efforts in understanding, and controlling, the infection process. Our molecular data also indicates that the Pyropia pathogen in New Zealand is genetically identical, or very similar, to samples growing on crops in the northern hemisphere (Korea, Japan). We believe that the molecular similarity with these markers, the grouping into a single species using several molecular species delimitation methods, including samples from Japan, plus the lack of robust characters to separate the two species indicates that they should be placed in taxonomic synonymy.
Pythium porphyrae M. Takah & M. Sasaki (1977). Transaction of the Mycological Society of Japan 18:279–285.
Heterotypic synonym: Pythium chondricola De Cock (1986). Mycotaxon 25:102, Fig. 1a–i.
The geographic range of Pythium porphyrae is now from the northern Atlantic to the northern and southern Pacific Oceans. This wide distribution, without any obvious biogeographic patterns and little or no genetic variation in standard molecular markers could indicate that this micro-organism, disperses easily and is truly ubiquitous (Fenchel and Finlay 2004). The host range is also varied, while infecting various genera and species of New Zealand Bangiales (Pyropia, Porphyra), we did not detect infections, other than on Pyropia plicata, in our population at Moa Point. It is possible that the virulence of Pythium porphyrae on other species is reduced, as the slower infection in culture suggests, and therefore not readily detected. Our isolate also did not infect filamentous gametophytic Bangiales (“Bangia”) nor any filamentous sporophytic phases (‘conchocelis’) which has been suggested before based on infection of Pythium marinum Sparrow (Kerwin et al. 1992), a species in clade B of Pythium (Lévesque and De Cock 2004). The cell wall composition of conchocelis phases is known to differ from the gametophytes (Mukai et al. 1981, Vreeland and Kloareg 2000), and carbohydrates are known to be important in spore attachment and penetration in P. porphyrae (Uppalapati and Fujita 2000). In our experiments, P. porphyrae was not able to infect members of the Florideophyceae, while the original collection of P. chondricola was from decaying red algae (e.g., Chrondrus crispus) but it is unclear if Pythium was a necrotroph or a saprotroph from the original descriptions (De Cock 1986). The full host range of this species in the field needs to be further investigated, while its host range in culture is known to be great, even infecting land plants (Klochkova et al. 2016a).
Our genetic species delimitation methods indicate that P. adhaerens may be conspecific with P. chondricola and P. porphyrae, although some species delimitation methods (ABGD, PTP) using ITS data indicate distinct species for P. adhaerens from P. chondricola–P. porphyrae. P. adhaerens was first described by Sparrow (1931) on the freshwater green alga Rhizoclonium hieroglyphicum (C. Agardh) Kützing, and it has been reported from land plants (sugar beet, maize, pea, tomato, and cucumber) (Sparrow 1932). Whether it is a distinct species, able to tolerate marine environments needs further study.
SUPPLEMENTARY MATERIAL
Supplementary Table S1. Detailed overview of hosts tested for specificity with Pythium porphyrae (http://www.e-algae.org).
Supplementary Table S2. Pythium samples used for ITS and COI trees from Genbank, with accession numbers (http://www.e-algae.org).
Supplementary Table S3. Pythium porphyrae collections from Wellington, New Zealand placed into culture, plus host and collection date (http://www.e-algae.org).
Supplementary Fig. S1. Maximum-likelihood phylogeny of all Bangiales from Sutherland et al. (2011) based on rbcL gene sequences. Samples used in infection with Pythium porphyrae experiments identified (marked with red boxes). Outgroups removed for clarity (http://www.e-algae.org).