Morphology and phylogenetic relationships of two Antarctic strains within the genera Carolibrandtia and Chlorella (Chlorellaceae, Trebouxiophyceae)
Article information
Abstract
The genera Carolibrandtia and Chlorella have been described as small green algae with spherical cell shapes that inhabit various environments. Species of these genera are often difficult to identify because of their simple morphology and high phenotypic plasticity. We investigated two small coccoid strains from Antarctica based on morphology, molecular phylogeny by two alignment methods which have been applied to previous phylogenetic studies of the genus Chlorella, and comparison of the secondary structures of nuclear small subunit (SSU) and internal transcribed spacer (ITS) rDNA sequences. Light microscopy of two strains revealed spherical cells containing chloroplasts with pyrenoids, and the morphological characteristics of the strains were nearly identical to those of other Chlorella species. However, based on the phylogenetic analyses of nuclear SSU and ITS rDNA sequences, it was determined that the Antarctic microalgal strains belonged to two genera, as the Chlorella and Carolibrandtia. In addition, the secondary structures of the SSU and ITS2 sequences were analyzed to detect compensatory base changes (CBCs) that were used to identify and describe the two strains. A unique CBC in the SSU rDNA gene was decisive for distinguishing strain CCAP 211/45. The ITS2 rDNA sequences for each strain were compared to those obtained previously from other closely related species. Following the comparison of morphological and molecular characteristics, we propose KSF0092 as a new species, Chlorella terrestris sp. nov., and the reassignment of the strain Chlorella antarctica CCAP 211/45 into Carolibrandtia antarctica comb. nov.
INTRODUCTION
The genus Chlorella Beijerinck, the spherical green alga, is one of the best-known microalgae worldwide. Since Chlorella vulgaris Beijerinck was described as the type species in 1890, over 100 species of Chlorella have been formally named. Chlorella species are often difficult to identify owing to their simple morphology and high phenotypic plasticity. Numerous studies have been conducted to find other criteria and methods for identifying those species (Huss et al. 1999, Krienitz et al. 2004, Luo et al. 2010, Heeg and Wolf 2015, Krivina and Temraleeva 2020). This genus consisted of only five “true” species based on biochemical and molecular data: Chlorella (Ch.) vulgaris, Ch. lobophora Andreyeva, Ch. sorokiniana Shihira and Krauss, Ch. heliozoae Pröschold and Darienko, and Ch. variabilis Shihira and Krauss (Huss et al. 1999, Krienitz et al. 2004, Pröschold et al. 2011). Bock et al. (2011) amended the generic description of Chlorella through a polyphasic approach, incorporating light microscopic observations, phylogenetic analyses based on small subunit (SSU) and internal transcribed spacer (ITS) rDNA aligned according to their secondary structures, and the compensatory base changes (CBC) within the secondary structure of ITS2. They also demonstrated that several strains, originally classified under the genus Dictyosphaerium, actually belong to Chlorella. The study recognized fourteen “true” Chlorella species.
Members of the genus Chlorella traditionally have been classified within the family Chlorellaceae, which is divided into two clades: the Chlorella clade and the Parachlorella clade (Krienitz et al. 2004, Luo et al. 2010). The Chlorella clade encompasses several genera, each with its own distinct morphology. These genera include Actinastrum, Didymogenes, Hegewaldia, Heynigia, Hindakia, Meyerella, and Micractinium (Luo et al. 2010, Pröschold et al. 2010). Several Chlorella-like taxa belonging to the Chlorella clade have been identified in subsequent studies, including Carolibrantdia ciliaticola (Hoshina and Nakada 2018, Hoshina et al. 2018), Meyerella similis (Krivina et al. 2022b), Meyerella krienitzii (Krivina et al. 2023c), Micractinium inermum (Hoshina and Fujiwara 2013), M. singularis, M. simplicissimum, M. variabile (Chae et al. 2019), M. tetrahymenae (Pröschold et al. 2020), M. kostikovii (Krivina et al. 2022a), M. lacustre, M. thermotolerans (Krivina et al. 2023a), Lewiniosphaera symbiontica (Pröschold and Darienko 2020), Neochlorella semenenkoi, and N. thermophila (Krivina et al. 2023b). These species are distinguished by their phylogenetic positions and unique CBCs in the nuclear SSU and ITS2 rDNA genes (Luo et al. 2006, 2010, Hoshina et al. 2018, Chae et al. 2019, Pröschold and Darienko 2020, Pröschold et al. 2020, Krivina et al. 2022a, 2022b, 2023a, 2023b, 2023c).
The genus Carolibrandtia (Ca.) was originally established as a monospecific genus with the type species Ca. ciliaticola (Hoshina, Kobayashi, Suzaki & Kusuoka) Hoshina & Nakada, which was discovered in the endocytic environment of some ciliate species (Hoshina and Nakada 2018, Hoshina et al. 2018). Carolibrandtia has a typical Chlorella-like morphology, yet it is distinct from other genera in the same family due to differences in the nucleotide sequence and secondary structure of its nuclear SSU rRNA. Within the Chlorella clade, only this genus has a C-G pair at the base of the E10_1 helix in the SSU (Hoshina et al. 2018).
The taxonomic classification of genera and species within the Chlorella clade has been established through molecular phylogeny and synapomorphic base changes in the SSU and ITS rDNA sequences, as outlined by Luo et al. (2010). Since this proposal, each genus within the Chlorella clade appears to form a well-defined monophyletic clade (Luo et al. 2010, Pröschold et al. 2010, 2020, Bock et al. 2011, Chae et al. 2019, Pröschold and Darienko 2020). However, recent phylogenetic studies have demonstrated that the polyphyly of the genus Chlorella (Krivina and Temraleeva 2020, Krivina et al. 2022a, 2022b, 2023a, 2023b, 2023c). The differences in the phylogenetic analysis results were due to variations in the alignment methods used for the datasets. Manual alignment based on the secondary structure of SSU and ITS indicated that the genus Chlorella formed a monophyletic clade, while analyses using multiple sequence alignment programs suggested a polyphyletic clade.
Morphologically simple Chlorella-like strains have been continuously reported in Antarctica despite their inaccessibility (Hu et al. 2008, Hodač et al. 2016, Chae et al. 2019, Martins et al. 2020, Kim et al. 2022). We examined two Chlorella-like Antarctic strains, CCAP 211/45 and KSF0092. The former was isolated by Sutton in 1970 and was identified as Chlorella antarctica (Fritsch) Wille based on its morphology. We isolated the other strain (KSF0092) from the surface of the bone in the South Shetland Islands, Antarctica. In this study, we investigated the morphological and molecular characteristics of two Chlorella-like strains from Antarctica, proposed a new combination, and described Chlorella terrestris sp. nov.
MATERIALS AND METHODS
Algal strains, culture conditions, and light microscopy
In this study, two Antarctic strains of the Chlorella clade were investigated (Table 1). The strain KSF0092 was collected in 2014 from the surface of the bone in Deception Island (62°58′36.50″ S, 60°40′31.90″ W), South Shetland Islands, Antarctica. The strain was isolated by transferring individual cells into a 6-well plate using a sterile Pasteur capillary pipette and deposited at the Korea Polar Research Institute (KOPRI) Culture Collections for Polar Microorganisms. Strain CCAP 211/45 was obtained from the Culture Collection of Algae and Protozoa (CCAP, UK). Two strains were cultivated on a Bold Modified Basal Freshwater nutrient solution B5282 (Sigma-Aldrich, St. Louis, MO, USA) at approximately 2°C with a light : dark cycle of 16 : 8 h. The illumination was provided by cool-white fluorescent lamps with 20–30 μmol photons m−2 s−1. The morphology of the strains was examined using an Axio Imager A2 microscope (Carl Zeiss, Oberkochen, Germany) with differential interference contrast optics (Fig. 1). Micrographs were obtained using an AxioCam HRC camera (Carl Zeiss). The morphological characteristics were described following the criteria established by Bock et al. (2011) and Hoshina et al. (2018).

Details of sampling information and GenBack accession numbers of small subunit (SSU) and internal transcribed spacer (ITS) rDNA sequences of strains used in this study
DNA extraction, polymerase chain reactions amplification, sequencing, and phylogenetic analyses
Genomic DNA from the strains in this study was extracted according to the manufacturer’s instructions for the i-genomic Plant DNA Extraction Kit (iNtRON Biotechnology, Seoul, Korea). Nuclear SSU, ITS1, 5.8S, and ITS2 rDNA were amplified by polymerase chain reactions (PCRs) using the primer pairs NS1 / NS4, NS5 / NS8, and ITS1 / ITS4 (White et al. 1990). Following amplification, the PCR products were purified by the manufacturer’s instructions using the MG PCR Product Purification Kit from Macrogen (Seoul, Korea) and sent to Macrogen for sequencing.
For phylogenetic analyses, we used the nuclear SSU, ITS1, 5.8S, and ITS2 rDNA sequences of the two strains and the sequences of 58 closely related chlorellacean strains. The GenBank accession numbers for all the sequences used in this study are shown in Fig. 2 and Supplementary Fig. S1. When introns were found in the SSU rDNA sequences, they were excluded from the alignment. Two datasets were generated based on the alignment methods: (1) a multiple alignment dataset (2,570 base positions) was performed on BioEdit 7.0.5.3 (Hall 1999) using the ClustalW algorithm, and (2) the other dataset (2,688 base positions) aligned according to its the secondary structures using the structure of Chlorella vulgaris (SAG 211-11b) (see Supplementary figs S1 & S2 in Luo et al. 2006). The evolutionary model for all datasets was determined using the program jModelTest 2 (Darriba et al. 2012) based on the Akaike Information Criterion. The GTR + I + G nucleotide substitution model was selected as the optimal model for Bayesian inference (BI) and Maximum likelihood (ML). BI was conducted using MrBayes 3.2.6 (Ronquist et al. 2012), employing four chains of Markov chain Monte Carlo iterations across 10,000,000 generations, with samples taken every 1,000 generations. The first 25% of trees were discarded as burn-in. ML analysis was performed using PhyML v3.0 (Guindon et al. 2010) with 1,000 bootstrap replicates. The phylogenetic tree data were visualized using FigTree v1.4.2 (available at http://tree.bio.ed.ac.uk/software/figtree/). To compare tree topologies, we consulted data from several studies, including Luo et al. (2010), Pröschold et al. (2010, 2020), Bock et al. (2011), Chae et al. (2019), Pröschold and Darienko (2020), as well as Krivina et al.’s series of studies in 2022 and 2023 (2022a, 2022b, 2023a, 2023b, 2023c).

Bayesian phylogenetic tree constructed from small subunit, internal transcribed spacer 1, 5.8S, and internal transcribed spacer 2 ribosomal DNA sequences. The phylogenetic tree shown was inferred based on the alignment according to the secondary structures. Numbers at each node are Bayesian probability (BP) (>0.95, left) and Maximum likelihood (ML) (>50%, right). * / * at each node indicates BP 1.00 and ML 100%. The new sequences are in bold font.
SSU and ITS2 secondary structures
To identify molecular signatures supporting the assigned species, the secondary structures of the E10_1 region of SSU rDNA and ITS2 were reconstructed. Subsequently, all sequences were manually compared with those from previous studies (Luo et al. 2006, Hoshina et al. 2018). Secondary structure models were produced by applying the minimum energy criterion using Mfold (Zuker 2003) and were visualized with PseudoViewer3 (Byun and Han 2009).
RESULTS
Morphological observations
The two Antarctic strains exhibited morphological characteristics common to Chlorella species, such as cell shape, chloroplasts, and pyrenoids (Table 2, Fig. 1). The vegetative cells of strain KSF0092 in culture were solitary, spherical or almost spherical shaped, and 3.4–7.2 μm in diameter without mucilaginous covering (Fig. 1A–D). The cells featured cup-shaped chloroplasts with spherical to ellipsoidal pyrenoids, which were encircled by several starch grains. Asexual reproduction was characterized by the formation of 2–8 autospores (Fig. 1E–H). Sexual reproduction and cell aggregation were not observed.
Cells of strain CCAP 211/45 were found to be solitary or colony, spherical or almost spherical shaped (3.8–5.7 μm) (Fig. 1I–M). Mucilage was not observed. The cells contained cup-shaped chloroplasts with spherical or ellipsoid pyrenoids. The pyrenoid was surrounded by several starch grains. Asexual reproduction occurred by the formation of 2–16 autospores (Fig. 1N–P). Sexual reproduction and cell aggregation were not observed.
Molecular phylogenetic analyses
The phylogenetic analyses of 60 SSU and ITS rDNA sequences demonstrated a relationship between the strains examined in this study and the Chlorellaceae (Fig. 2, Supplementary Fig. S1). To verify that the two Antarctic strains formed separate subclades with each type species of the genera Chlorella and Carolibrandtia, two distinct alignment methods were employed. Using the first method, which aligned the dataset according to the secondary structures, two lineages were revealed: the Chlorella and Parachlorella clades (Fig. 2). The phylogenetic tree revealed ten lineages within Chlorella clade. The support for the Chlorella subclade is moderate to weak in both BI and ML. Strain KSF0092 was placed within the Chlorella subclade and was sister to Ch. pituita C. Bock, Krienitz & Pröschold. This strain belongs to the same clade as strain WT-3-1-L2 (Chlorella cf. pituita) (Sommer et al. 2020) but differs by 11 nucleotide substitutions. The Carolibrandtia subclade was strongly supported in the ML bootstrap and Bayesian analyses and was clearly separated from the Chlorella subclade. Within the Carolibrandtia subclade, strain CCAP 211/45 was closely related to Ca. ciliaticola, the type species of the genus (Fig. 2). The genetic differences between strain CCAP 211/45 and Ca. ciliaticola varied, ranging from 48 to 50 nucleotide substitutions. In the second method, where the dataset was aligned using the ClustalW algorithm, the strain KSF0092 was placed within a strongly supported subclade (BI / ML = 1.00 / 100) containing the Chlorella type species, Ch. vulgaris (Supplementary Fig. S1). Chlorella pituita was determined to be the species most closely related to KSF0092, differing from it by 81 nucleotide substitutions. The genus Carolibrandtia is strongly supported in both ML and BI. The Carolibrandtia subclade comprises Ca. ciliaticola and strain CCAP 211/45. Notably, Ca. ciliaticola (NIES 4033) differs from strain CCAP 211/45 by 45 nucleotide substitutions.
Secondary structures of SSU and ITS2
The general secondary structures of CCAP 211/45 and KSF0092 are presented in Figs 3 & 4, and they show similarities to those found in the members of Chlorella and other genera of the Chlorella clade. The secondary structures of the 10 to E10_1 helices of the SSU rDNA for both KSF0092 and CCAP 211/45 are shown in Fig. 3. At the base of the E10_1 helix, strain KSF0092 has a U-A pair, whereas strain CCAP 211/45 has a C-G pair. Strain CCAP 211/45 had identical SSU rDNA sequences and belonged to the same genus, Carolibrandtia.

Secondary structure diagrams for small subunit ribosomal DNA from 10 to E10_1 helices, following the naming system of Luo et al. (2006). The key compensatory base change at the base of E10_1 helix that distinguishes Carolibrandtia from Chlorella is shown as a black circle.
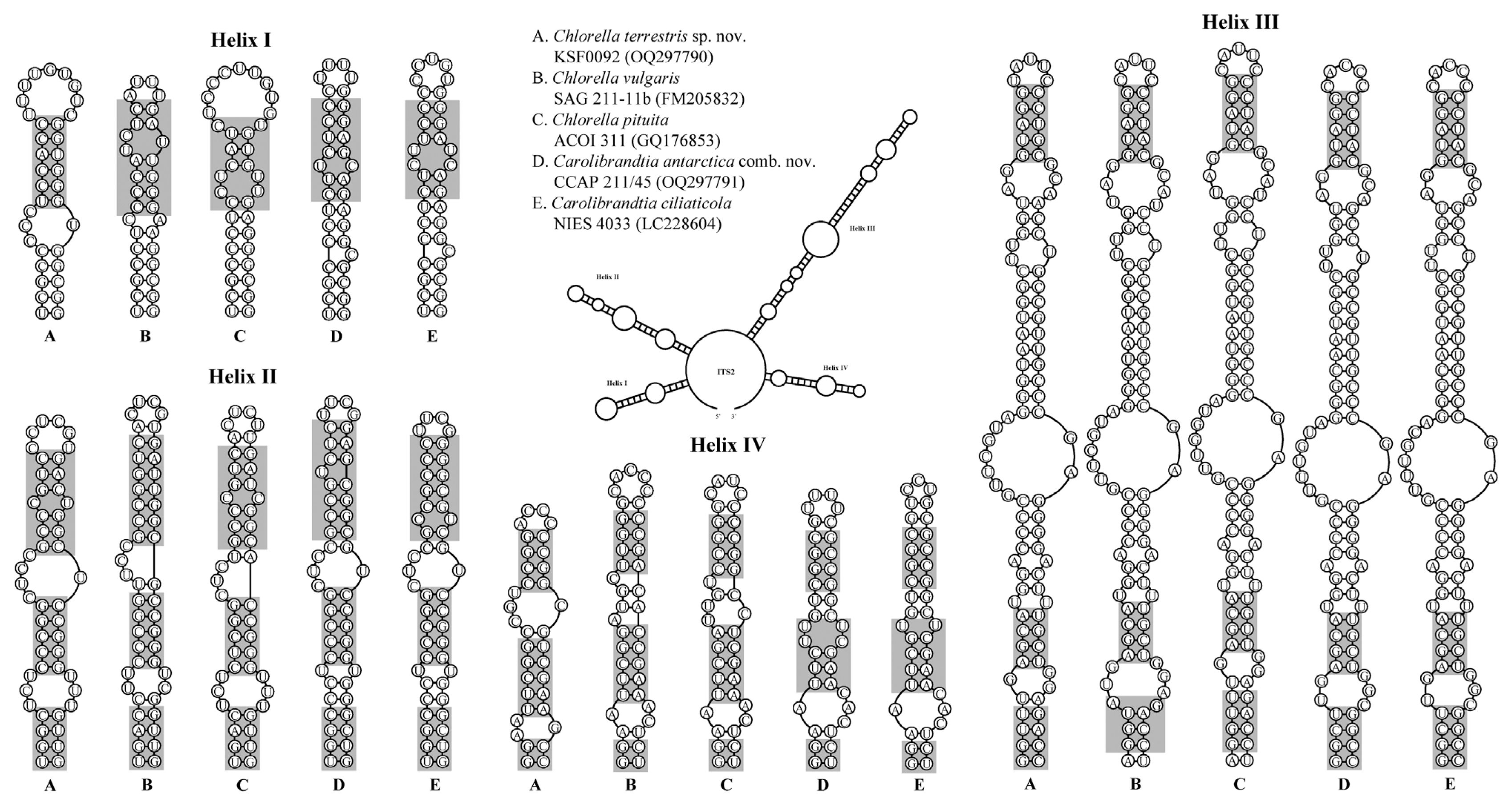
Secondary structure of internal transcribed spacer 2 rDNA sequences among the species of the Chlorella clade. The compensatory base changes (CBCs) and hemi-CBCs are marked by grey boxes.
Fig. 4 shows four typical helices called helices I–IV for ITS2 according to Mai and Coleman (1997). The differences among species in ITS2 secondary structure revealed that each species could be identified based on distinctive CBCs and structures. The differences in base pairs within the conserved ITS2 region among the genera Chlorella and Carolibrandtia are summarized in Fig. 5. The predicted secondary structure of the nuclear rDNA ITS2 of KSF0092 was compared to that of other species of Chlorella (Fig. 4). Strain KSF0092 differed from Ch. vulgaris (SAG 211-11b) with two CBCs and seven hemi-CBCs in helices I–IV. Compared to Ch. pituita, nine hemi-CBC sites of the ITS2 secondary structures were identified in KSF0092 (Fig. 5). Among Carolibrandtia, the strains showed little variation within the conserved region of ITS2. Strain CCAP 211/45 is closely related to strain NIES 4033, which is an authentic strain of Ca. ciliaticola, differing by one CBC in Helix II of ITS2 (Fig. 5).

Comparison of internal transcribed spacer 2 secondary structure of two Antarctic strains with other Carolibrandtia and Chlorella species. Hyphens (−) and dots (...) indicate gaps and omitted regions, respectively. The positions marked by empty boxes indicate the presence of compensatory base changes (CBCs) and hemi-CBCs compared to other species. Numbers of CBCs and hemi-CBCs are indicated on the right.
Taxonomical conclusions
Chlorella terrestris H. Chae, H.-G. Choi and Ji Hee Kim sp. nov. (Fig. 1A–H)
Description. Cells solitary, spherical, or almost spherical, and 3.4–7.2 μm in size. Absence of mucilage. Single chloroplast, parietal, cup-shaped, with a spherical to ellipsoidal pyrenoid. The pyrenoids were encircled with two starch grains. Asexual reproduction by formation of 2–8 autospores. Sexual reproduction was unknown and cell aggregates were not observed. This species differs from other species in its genus due to variations in the nucleotide sequences of ITS2 and distinct barcoding signatures.
Type locality. Deception Island (62°58′36.50″ S, 60°40′ 31.90″ W), South Shetland Islands, Antarctica.
Holotype. Strain KSF0092, cryopreserved and deposited at KOPRI, Incheon, Korea.
Etymology. The specific epithet refers to the terrestrial habitat from which the new species was collected.
GenBank accession number. OQ297790.
Carolibrandtia antarctica (Fritsch) H. Chae, H.-G. Choi & Ji Hee Kim comb. nov. (Fig. 1I–P)
Basionym. Chlorosphaera antarctica Fritsch 1912, Freshwater algae. In: National Antarctic Expedition 1901–1904 Natural History. Vol. VI. Zoology and Botany [Part 3]. (Anon. Eds), pp. 1–66. London: Printed by the Trustees of the British Museum.
Homotypic synonym. Chlorella antarctica (Fritsch) Wille 1924, Süsswasseralgen von der deutschen Südpolar-Expedition auf dem Schiff “Gauss”. In: Deutsche Sudpolar-Expedition 1901–1903. Bd. VIII. (Drygalski, E. von Eds), pp. 373–444.
Holotype. Figs 2–6, Fritsch 1912.
Epitype (designated here to support the holotype). Material of the strain CCAP 211/45 was cryopreserved in a metabolically inactive state at the Culture Collection of Algae and Protozoa (CCAP) in Oban, Scotland.
Emended description. Cells solitary or colonial, spherical or almost spherical, and 3.8–5.7 μm. Absence of mucilage. Single chloroplast, parietal, cup-shaped, with a spherical to ellipsoid pyrenoid. The pyrenoids were covered with starch grains. Asexual reproduction by formation of 2–16 autospores. Sexual reproduction was unknown and cell aggregates were not observed. This species differs from others in its genus due to variations in nucleotide sequences of ITS2 and distinct barcoding signatures.
GenBank accession number. OQ297791.
DISCUSSION
The traditional classification of the family Chlorellaceae, based on morphological characteristics, is inconsistent with phylogenetic analyses using molecular marker genes, reflecting the family’s high phenotypic plasticity and the instability of its morphological traits. New genera and species within the Chlorella clade have been identified and described based on molecular phylogeny and the secondary structures of SSU and ITS rDNA sequences (Luo et al. 2010, Bock et al. 2011, Hoshina and Fujiwara 2013, Hoshina et al. 2018, Chae et al. 2019, Pröschold et al. 2020, Krivina et al. 2022a, 2022b, 2023a, 2023b, 2023c). Bock et al. (2011) revised the species concept of the genus Chlorella based on a polyphasic approach, emphasizing the use of molecular signatures to distinguish the Chlorella species. However, Krivina and Temraleeva (2020) confirmed the polyphyly of the genus Chlorella and, the CBC approach was ineffective for Chlorella species. The differences in alignment methods were identified as a factor influencing the results of the two phylogenetic studies. We aligned two datasets using the methods described by Bock et al. (2011) and Krivina and Temraleeva (2020), respectively.
The newly isolated Chlorella terrestris sp. nov. (KSF0092) clustered with the German strain WT-3-1-L2 (Sommer et al. 2020) within the same clade and was distinguished from other members of the genus Chlorella by SSU and ITS rDNA, regardless of the alignment methods of sequences (Fig. 2, Supplementary Fig. S1). Chlorella terrestris exhibited morphological characteristics similar to those of the Chlorella type species, Ch. vulgaris; however, a cell size of 1.5–10 (–13.3) μm (Tsarenko and John 2011), which is a broader range of size than strain KSF0092 cells (3.4–7.2 μm), all other characters matched to the latter (Table 2). Molecular analyses supported the identification of Ch. pituita, which was the closest relative to Ch. terrestris (Fig. 2, Supplementary Fig. S1). The two Chlorella species are distinguished by their life forms and the presence or absence of a gelatinous envelope (Bock et al. 2011). As shown in Figs 4 & 5, Ch. terrestris and two closely related species could be clearly distinguished by CBCs and hemi-CBCs in their ITS2 secondary structures.
Most of the known species of the genus Chlorella, except for the “true” species, have been described based on microscopical investigations. Over time, numerous studies have been conducted to revise the systematics of this genus, and some have focused on physiological data (Shihira and Krauss 1965, Kessler and Huss 1992). However, the comparison and judgement of morphological and physiological characteristics are quite difficult. For example, Shihira and Krauss (1965) proposed Ch. photophila based on its nutritional requirements. Morphologically, this species is very similar to Ch. vulgaris (Komárek and Fott 1983) (Table 2). One strain, designated Cambridge Collection No. 211/8a, was likely identical to strain UTEX 26. This strain was later transferred to Ch. vulgaris (Kessler and Huss 1992). Therefore, it is possible that both species represent only one species, Ch. vulgaris, and further investigation is needed.
After Chlorella antarctica was first reported as a species in the genus Chlorosphaera by Fritsch in 1912, reproduction through autospores was observed and transferred to the genus Chlorella (Wille 1924). The strain CCAP 211/45, previously identified as Ch. antarctica, is morphologically suitable for the traditional circumscription of the genus Chlorella (Fig. 1I & J). However, phylogenetic analysis has revealed the formation of the same clade as Ca. ciliaticola, a type species of the genus Carolibrandtia (Fig. 2, Supplementary Fig. S1). Carolibrandtia ciliaticola was similar to the morphological characteristics of strain CCAP 211/45; however, the strain often showed colonization and was smaller than Ca. ciliaticola. The CBC at the base of E10_1 helix in SSU (C-G in Carolibrandtia vs. U-A in Chlorella), the molecular signature described by Hoshina et al. (2018), was also observed in strain CCAP 211/45 (Fig. 3). Based on the strain CCAP 211/45, Ch. antarctica was transferred to the genus Carolibrnadtia.
In the taxonomic research of morphologically simple Chlorella-like strains or species, crucial characters at the genus or species level have been derived from the secondary structure of SSU and ITS rDNA sequences, as evidenced by several studies (Luo et al. 2010, Pröschold et al. 2010, 2020, Bock et al. 2011, Chae et al. 2019, Pröschold and Darienko 2020, Krivina et al. 2022a, 2022b, 2023a, 2023b, 2023c). Notably, the alignment of sequences based on their secondary structure has different results compared to using a multiple sequences program (Fig. 2, Supplementary Fig. S1). For example, depending on the alignment method, the genus Chlorella exhibited either a monophyletic or polyphyletic clade (Bock et al. 2011, Krivina and Temraleeva 2020). Through phylogenetic analyses with two different alignment methods, our study was able to distinguish Ch. terrestris from other known “true” Chlorella species supporting its recognition as a “true” Chlorella species. However, in the case of the genus Neochlorella, the phylogenetic analyses based on secondary structures suggest it belongs to the Chlorella subclade (Fig. 2). Furthermore, comparisons of SSU and ITS2 secondary structures failed to reveal unique base pairs that distinguish Neochlorella from the genus Chlorella. Consequently, additional taxonomic studies on this genus are necessary for a more comprehensive understanding.
The genera Carolibrandtia and Chlorella have similar distributions in terrestrial habitats and symbioses. The genus Chlorella contains a relatively large number of species and has been reported not only in terrestrial and symbiosis, but also in marine environments (Bock et al. 2011, Pröschold et al. 2011, Krienitz and Bock 2012, Hodač et al. 2016, Darienko et al. 2019). Interestingly, according to the phylogenetic analyses, some marine strains belong to Ch. vulgaris; however, these strains showed different morphological characteristics under marine and freshwater conditions (Darienko et al. 2019). The high phenotypic plasticity of morphological characteristics according to the environment represents a difficulty in the identification and classification of species consisting only of previously identified morphological characteristics. Microalgae living in Antarctica, a low-temperature environment, are difficult to classify solely based on their morphological characteristics. Here, we report a new species and a combination of Antarctic strains, indicating that Antarctic biodiversity is richer than expected. Further taxonomic studies on Antarctic green coccoid strains could improve our understanding of the ecological speciation of green coccoid microalgae.
ACKNOWLEDGEMENTS
This work was supported by the Korea Polar Research Institute (KOPRI) grant funded by the Ministry of Oceans and Fisheries (KOPRI PE23130 and PE23140).
Notes
The authors declare that they have no potential conflicts of interest.
SUPPLEMENTARY MATERIALS
Bayesian phylogenetic tree constructed from small subunit, internal transcribed spacer 1, 5.8S, and internal transcribed spacer 2 ribosomal DNA sequences (https://www.e-algae.org).

