Synchronous nuclear division and the role of the cytoskeleton in the multinucleate red alga Griffithsia monilis
Article information
Abstract
Most taxonomic groups of organisms harbor temporarily or permanently multinucleate cells in all or parts of their bodies. Each nucleus in the same cytoplasm responds almost identically to environmental cues, but little is known about the signals that mediate their coordinated division. In this study, we used Griffithsia monilis, a multinucleated giant cell, to investigate how its nuclear division occurs and the role of cytoskeleton in this process. Our results show that nuclear division is exquisitely coordinated and synchronized, but that nuclear division and chloroplast division are not coupled to each other. Microtubules are known to play an important role in synchronized nuclear division in some large multinucleate green algae, and microtubule arrangement is involved in shaping the cytoplasmic domains of each nucleus. However, we found no evidence for the involvement of the cytoskeleton in the synchronized nuclear division or regular nuclear arrangement in G. monilis. Although the nuclei were arranged at very regular intervals, these intervals became irregular during nuclear division, and there was no regular arrangement of actin or microtubules to maintain the spacing between the nuclei. Neither cortical microtubules nor spindle microtubules were physically connected to other neighboring nuclei during nuclear division, suggesting that microtubules are not involved in the coordination of nuclear division in G. monilis.
INTRODUCTION
Mitosis is essential for growth, development, and repair of multicellular organisms and is tightly controlled by multiple regulators and the checkpoint machinery (Nigg 2001, Storchova and Pellman 2004). Multinucleate states have evolved many times in unrelated plant species including 60 families of land plants and in diverse algal lineages including red algae (Niklas et al. 2013). Multinucleate cells pose a challenge to the coordination and regulation of mitosis, as each nucleus can have a different cell cycle status and respond differently to environmental cues (Yabuta et al. 2007). Many multinucleated cells ensure that all their nuclei divide together by synchronizing the mitotic events of all the nuclei within a cell, so that they enter and exit mitosis simultaneously. This is known as synchronous mitosis, and it is observed in some fungi, algae, and animal cells that have multiple nuclei (Lüttke and Bonotto 1982, Menzel 1994, Gladfelter 2006, Simon et al. 2021). However, not all multinucleate cells undergo synchronous mitosis. Some cells, such as the filamentous fungus Ashbya gossypii, undergo asynchronous mitosis, in which each nucleus divides independently in time and space (Jorgensen et al. 2007). Asynchronous mitosis may have some advantages, such as allowing flexibility and adaptation to environmental changes and generating genetic diversity (Gladfelter 2006).
Synchronous mitosis may have more critical advantages for multinucleated cells, such as maintaining genomic stability, avoiding nuclear competition, and facilitating cell differentiation, but it also requires a complex mechanism to coordinate the cell cycle progression of all the nuclei within a cell (Simon et al. 2021). Parasitic infections or genetic abnormalities can sometimes turn uninucleate cells into multinucleate ones: plants can form giant cells with up to 100 nuclei when infected by nematodes (Favery et al. 2016). The unicellular marine organism Sphaeroforma arctica, closely related to multicellular animals, is an organism with a Plasmodium-associated mode of proliferation that produces cells of up to 128 nuclei (Ondracka et al. 2018). To understand Plasmodium-associated proliferation, it is essential to study the synchronization process in multinucleated cells.
Microtubules, a cytoskeletal structure, have been reported to mediate synchronous mitosis in multinucleate cells by acting as an inter-nuclear signaling pathway in some animals (Meissner and Jaenisch 2006). Epifluorescence microscopy studies have reported that microtubules are also involved in synchronized nuclear division in some large multinucleate green algae, and this microtubule array is thought to play an important role in shaping the cytoplasmic region of each nucleus (e.g., Goff and Coleman 1987, McNaughton and Goff 1990). Early studies of the cell division and the role of the cytoskeleton in red algae were made using electron microscopy (e.g., Pueschel 1990) and studies using fluorescent probes focused mainly on microfilaments (Garbary et al. 1992, McDonald et al. 1992, 1993). The role of the cytoskeleton in other cellular responses is even less understood. Studies of the function of the cytoskeletal component in red algae have mainly been carried out during fertilization (Shim et al. 2020), as it is relatively easy for inhibitors to enter the reproductive cells. Actin has been shown to mediate the movement of spermatial nuclei within the trichogynes of several red algae during fertilization (Kim and Kim 1999, Wilson et al. 2002, Shim et al. 2021). However, investigations on the involvement of microtubules in various cellular responses in red algae have only recently begun (Moon et al. 2022).
Little is known about how nuclear division occurs and the role of the cytoskeleton in this process in red algal multinucleate cells. In this study, we have developed a staining method that allows us to observe the cytoskeleton, in particular microtubules, during nuclear division, and we will discuss how the cytoskeleton is involved in mitosis in Griffithsia monilis.
MATERIALS AND METHODS
Algae cultures
A gametophytic strain of Griffithsia monilis was provided by Prof. J. A. West in 2008 and kept in laboratory culture. Thalli were maintained in unialgal cultures in IMR medium (Klochkova et al. 2005) at 20°C on a 16 h light / 8 h dark cycle with illumination of >20 μmol photons m−2 s−1 provided by cool white fluorescent lighting. Plants were transferred to fresh IMR medium every 1–2 weeks.
Actin staining
To visualize the actin cytoskeleton, the filaments were fixed for 30 min in 3.7% (w/v) in formaldehyde diluted with EMP buffer (10 mM ethyleneglycol-bis[β-aminoethylether]-N,N′-tetraacetic acid [EGTA], 5mM MgSO4, 20 mM piperazine-N,N′-bis[2-ethanesulfonic acid] [PIPES]-KOH, pH 7.0) in 2% NaCl (w/v) (Yoshida and Shimmen 2009). After fixation, the cells were cut with a razor blade and were rinsed three times with phosphate-buffered saline (PBS; 8 mM Na2HPO4, 2 mM NaH2PO4, 140 mM NaCl; pH 7.4), and then placed in 0.5% (v/v) Triton X-100 diluted in EMP. The cells were then washed again three times with PBS before incubation in a staining solution of FITC-phalloidin (Sigma-Aldrich, St. Louis, MO, USA) (Wieland et al. 1983) for 12 h at 4°C in the dark. FITC-phalloidin was prepared as a stock solution of 50 μg mL−1 in methanol and was stored at −20°C in the dark. The stock was diluted with EMP to a final concentration of 2.5 μg mL−1. An IX73 inverted microscope (Olympus, Tokyo, Japan) equipped with fluorescence devices and a TCS SP5 confocal microscope (Leica Camera AG, Wetzlar, Germany) were used to observe the stained cells.
Microtubule staining
The filaments were fixed for 30 min in 3.7% formaldehyde in EMP containing 2% NaCl. After fixation, the cells were cut open with a razor blade and were rinsed three times in PBS buffer and then placed in 0.5% Triton X-100 for 30 min. The filaments were again rinsed three times with PBS buffer. For microtubule staining, the cells were incubated overnight at 4°C in the dark with the rat monoclonal anti-tubulin antibody conjugated to Alexa Fluor 488 (Abcam, Cambridge, UK), diluted at a 1 : 100 ratio in EMP.
Nuclear staining
Hoechst 33342 (Sigma-Aldrich) was used for nuclear staining. The stock solution of the reagent was prepared at a concentration of 1 mg mL−1 in sterile distilled water. Filaments were fixed as described above for microtubules. The cut cells were washed three times with seawater and then placed in Hoechst 33342 diluted at 1/1,000 in seawater for 30 min. The cells were then washed three times with fresh seawater, and the nuclei were observed under a fluorescence microscope with a UV filter.
RESULTS
Cell structure of Griffithsia monilis
The branched filaments of G. monilis is composed of dozens of multinucleate cells, with the largest cells near the base and cells getting smaller in both directions (Fig. 1A). When G. monilis cells were examined under a confocal microscope, a large vacuole filled almost the entire interior space, and organelles such as chloroplasts and nuclei were distributed in the thin layer between the cell membrane and the vacuole membrane (Fig. 1B, yellow arrow). Depending on the size of the cell, ~500–2,000 nuclei are evenly distributed in the cytoplasm (Fig. 1C & D). Chloroplasts are scattered throughout the cytoplasm, but their number per nucleus is variable and their distance from the nucleus is not constant (Fig. 1E). Fluorescence staining revealed a regular spatial arrangement of nuclei and a large number of microfilaments between nuclei, and there is variation in staining intensity (Fig. 1D–F). However, there was no obvious connection between the microfilaments surrounding each nucleus (Fig. 1F). Prior to nuclear division, microtubules were densely distributed beneath the plasma membrane surrounding the entire cell, with fine microtubules branching off from a network of coarser microtubules (Fig. 1I).
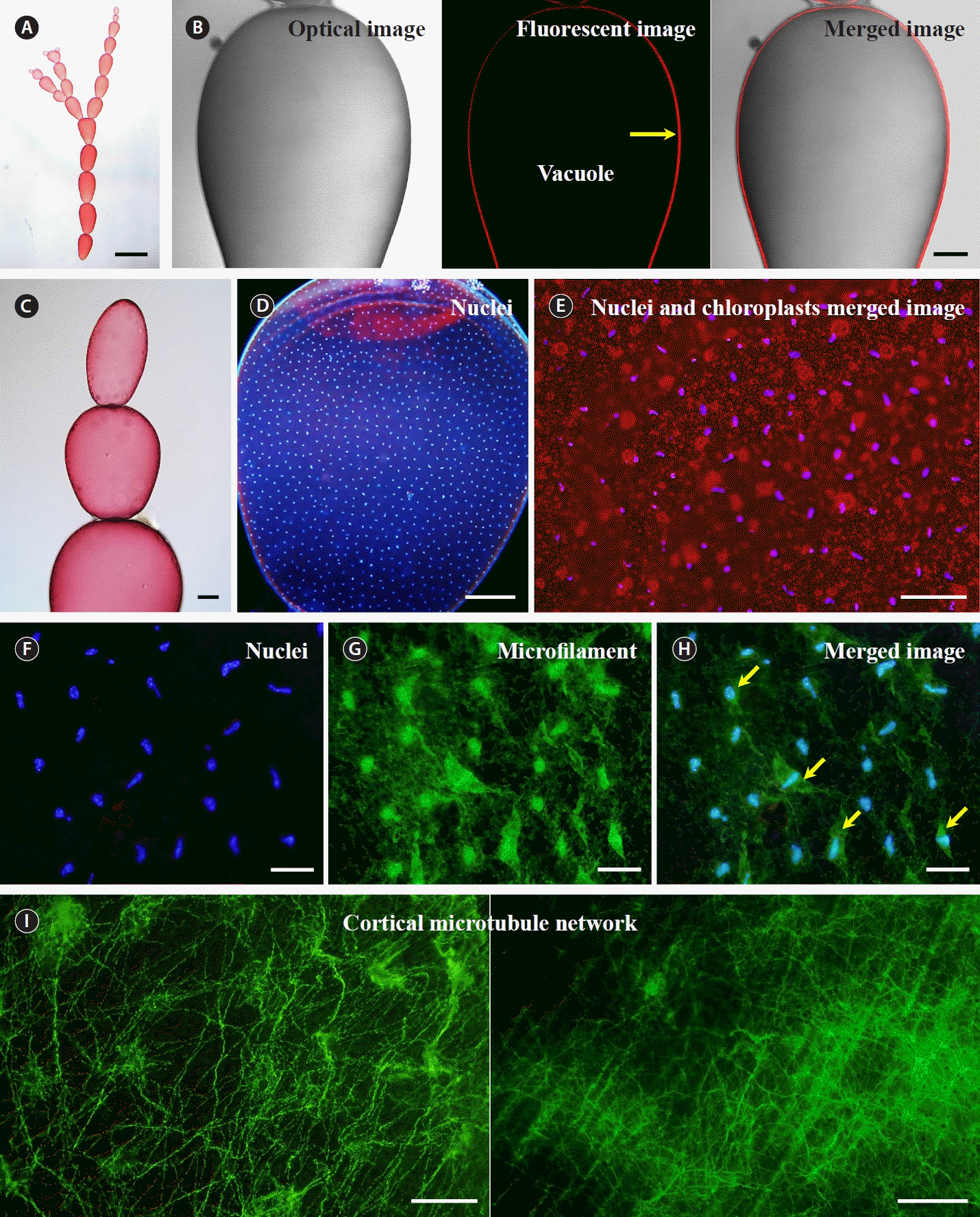
Cell structure of Griffithsia monilis. (A) Branched filamentous body. (B) From left to right, optical image, fluorescence image, and merged image under a confocal microscope. Yellow arrow, cytoplasmic layer based on autofluorescence of chloroplasts. (C) Apical end of filamentous body. (D) Image of regularly spaced and distributed nuclei. (E) Image of nuclear staining superimposed on chloroplast autofluorescence. (F–H) From left to right. (F) Nuclei. (G) Microfilaments. (H) Merged image of nuclei and microfilament staining. Yellow arrows, nuclei with few microfilaments around them. (I) Microtubule network. Scale bars represent: A, 2 mm; B–D, 200 μm; E, 50 μm; F–I, 20 μm.
Microtubules during mitosis
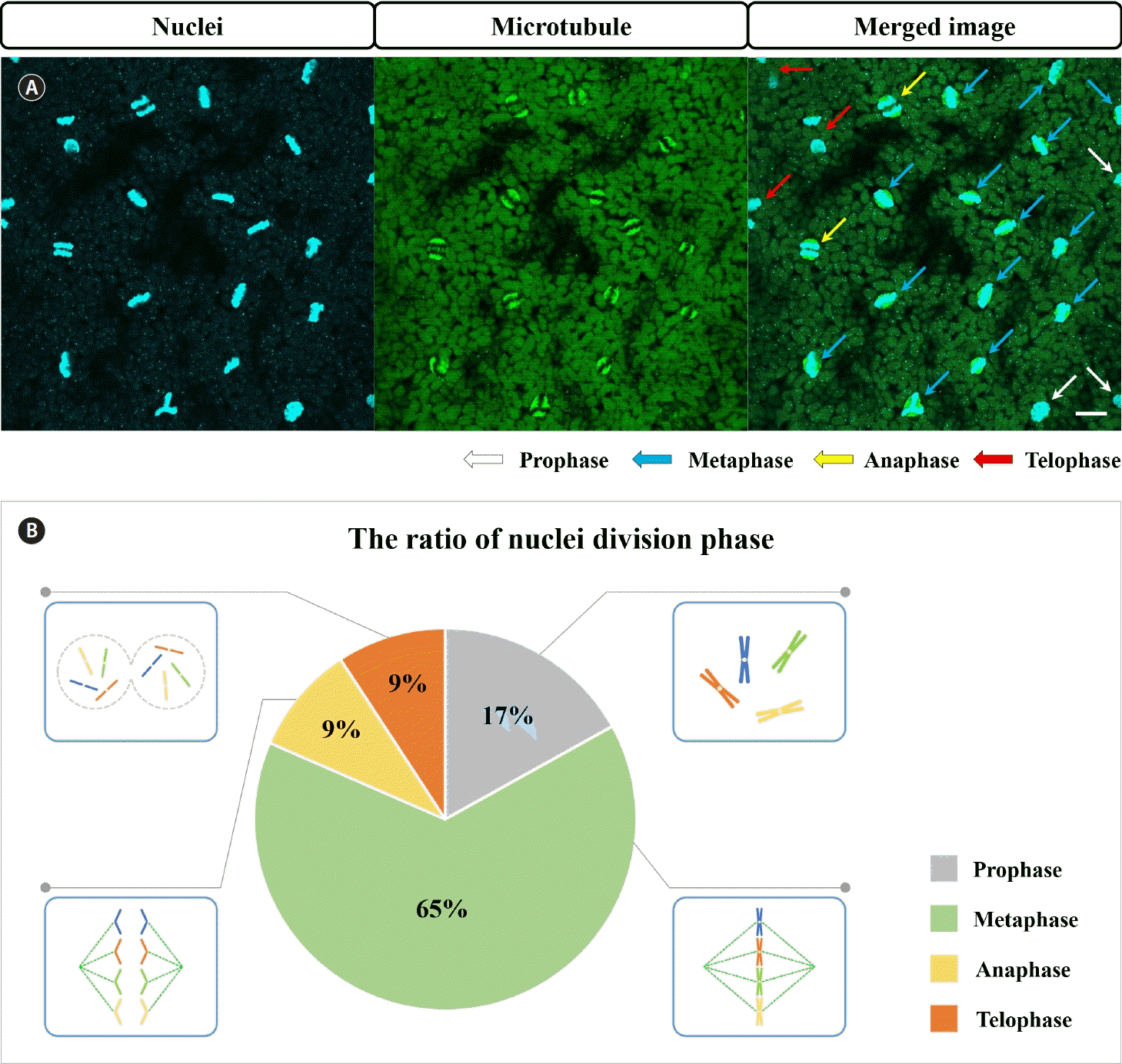
When we performed nuclear staining at different times of the day, we observed that mitosis in G. monilis occurs in a synchronized manner only during the night (Fig. 2A). During mitosis, microtubules were only observed around the nuclei, with no microtubules visible elsewhere, including around chloroplasts, which are located close to the nucleus (Fig. 2B). Based on the size of the chloroplasts, it appears that chloroplasts rarely divide during mitosis (Fig. 2B & C). Spindle microtubules are clearly visible (Fig. 2C, white arrows). When the chromosomes finished moving towards the poles, the microtubules disassembled and eventually disappeared (Fig. 2C, red arrow). When focusing near the cell surface at the time of nuclear division, the cortical microtubule network was not as pronounced as before, and the microtubules had a distinct radial extension from the poles (Fig. 2D). At the time of nuclear division, the distance between each nucleus is slightly irregular, and spindle microtubules extending radially from the poles do not appear to be connected to microtubules from other nuclei (Fig. 2D). Nuclear division was exquisitely synchronized, with 65% of mitosis in metaphase simultaneously (Fig. 3). None of the nuclei observed were copying DNA (synthesis or S phase) or preparing to divide (gap 2 or G2 phase); all were in mitosis, with 65% of them in metaphase (Fig. 3B).
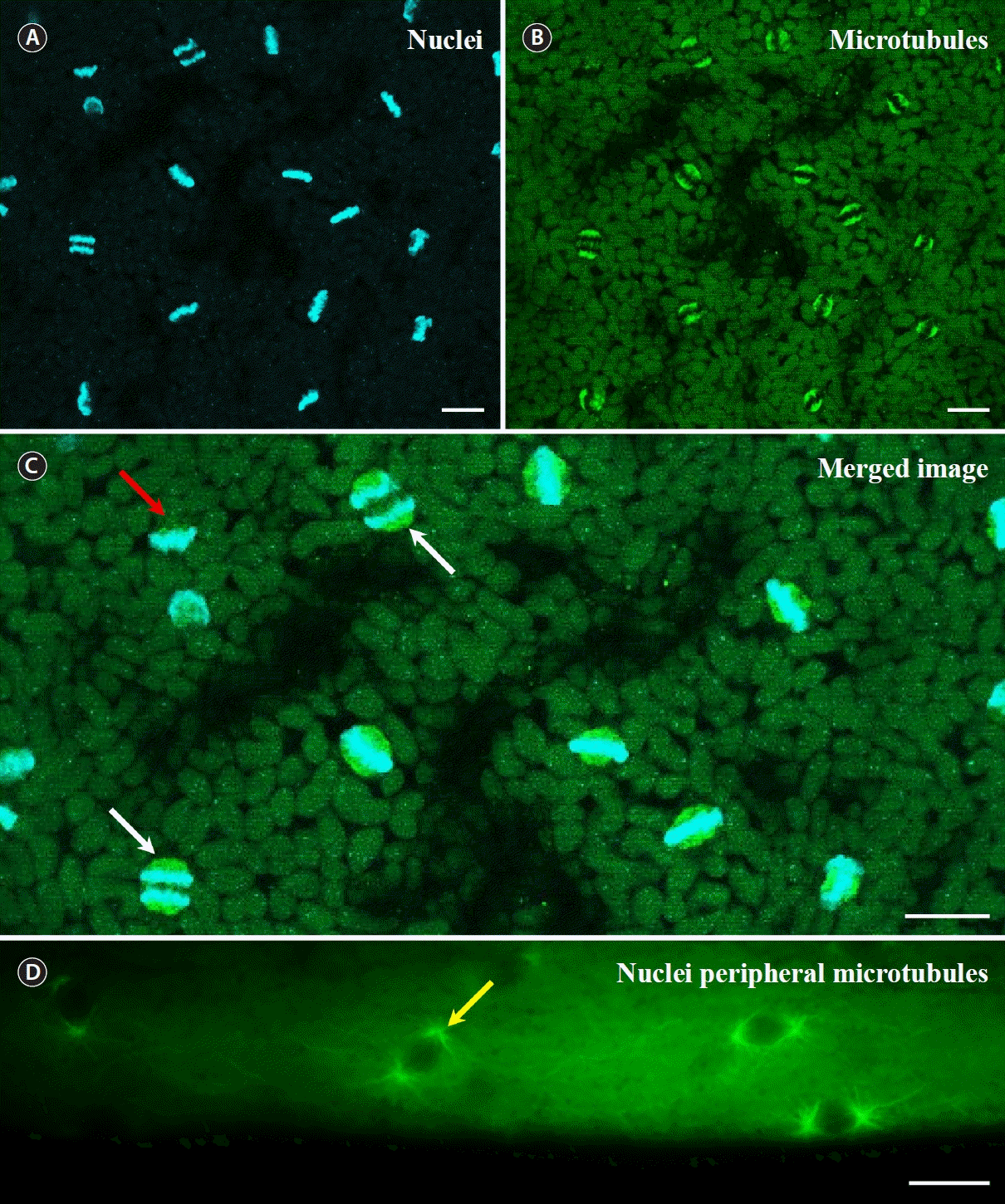
Nuclei and microtubules of Griffithsia monilis during mitosis. (A) Chromosomes. (B) Microtubules and spindles. (C) Merged image of A and B. White arrows, microtubule spindles that pull chromosomes towards the poles; red arrow, microtubules disassembled during anaphase. (D) Microtubules during mitosis in nuclei located close to the cell surface. Yellow arrow, microtubules extending radially from the pole. Scale bars represent: A–D, 10 μm.
DISCUSSION
Multinucleated cells are found in most taxonomic groups of organisms. Many fungi, algae, plants, and animals consist entirely or partly of multinucleated cells and sometimes some or all of the tissues of a unicellular organism become temporarily multinucleated (Niklas et al. 2013). These phenomena show that nuclear division is not inseparable from cell division (as Strasburger noted in 1913). Cells that have become multinucleated, with nuclear division separated from cell division, face another challenge. All nuclei in the same cytoplasm respond in a similar way to an environmental stimulus, but for a stimulus localized to a small part of a large cell, such as a G. monilis cell, it is a very interesting question whether all or only some of the nuclei in the cell will transcribe responsive genes. The wound-healing process in G. monilis provides a clue to this question (Moon et al. 2022). When a cell dies in the G. monilis filament, neighboring cells produce repair cells, and during the process of wound healing by somatic fusion between them, the repair cells produce reactive oxygen species (ROS) only on the facing sides (Moon et al. 2022). Just because ROS production occurs in the localized region of the cell does not mean that only nuclei in the vicinity are involved in this signaling, but it does mean that these nuclei are exposed to different stimuli than nuclei in other parts of the same cell. Our results show that nuclear division in G. monilis is exquisitely coordinated. So what are the signals that enable this coordination?
One hypothesis for the molecular basis of synchronous mitosis in multinucleate cells is that cell cycle regulators shuttle between nuclear and cytoplasmic compartments, allowing them to diffuse and coordinate multiple nuclei in a common cytoplasm. For example, in the multinucleate slime mold Physarum polycephalum, cyclin B1, a key protein that controls the entry into mitosis, has been shown to shuttle between the nucleus and the cytoplasm during interphase, and to accumulate in all nuclei prior to mitosis (Gladfelter 2006). Similarly, in some algae that have hundreds of nuclei in a single cell, such as Caulerpa, it has been suggested that calcium ions act as secondary messengers to synchronize the nuclear division (Menzel 1994). Another example is that microtubules mediate synchronous mitosis in multinucleated cells by acting as a conduit for inter-nuclear signaling. In some animal cell types that have two or more nuclei per cell, such as syncytiotrophoblasts and cardiomyocytes, it has been observed that microtubules connect adjacent nuclei and form a common spindle during mitosis, allowing the exchange of molecular factors or mechanical forces between nuclei to coordinate their cell cycle phases (Meissner and Jaenisch 2006). Our results show that microtubules or spindles are not physically connected to other neighboring nuclei or spindles during nuclear division in G. monilis, suggesting that coordination is mediated by a different signal.
Chloroplast and mitochondrial replication are not intrinsically linked to nuclear or cytoplasmic division (Imoto et al. 2011). Our results also showed that nuclear division and chloroplast division are not coupled in G. monilis. In addition to nuclear division, the division of subcellular organelles such as chloroplasts and mitochondria has been reported to be mediated by ROS signaling (Shapiguzov et al. 2012). It is possible that the synchronization of nuclear division in G. monilis is regulated by Ca2+-mediated ROS signaling: a study of the wound-healing process in the same species showed that the hydrogen peroxide production induced by the intracellular calcium influx mediates wound healing, including the division of repair cells (Moon et al. 2022). The same type of signaling has also been reported to mediate the fertilization process in other red algae (Shim et al. 2021). Further studies are needed to determine whether Ca2+-mediated ROS signaling is also involved in the coordination of nuclear division in multinucleate cells.
In uninucleate cells, cytokinesis follows karyokinesis, and restores a specific nucleus-to-cytoplasm ratio. In multinucleate cells, cytokinesis is absent or rare; no plasmalemma boundary defines the cytoplasmic territory of a single nucleus (Goff and Coleman 1990). Several genera of large multinucleate green algae have been studied by epifluorescence light microscopy and have shown that patterns of cytoplasmic organization are related to nuclear cytoplasmic domains (McNaughton and Goff 1990). They showed that regularly spaced nuclei undergo synchronous divisions in the stationary cytoplasm of these green algae, and that microtubules radiate from the regularly spaced nuclei in late telophase or early interphase, and that these microtubule arrays play a role in establishing the cytoplasmic “domain” of each nucleus (McNaughton and Goff 1990). A similar pattern of cytoplasmic organization, related to nuclear cytoplasmic “domains”, has been reported in G. monilis using epifluorescence light microscopy (Goff and Coleman 1987). Our results show that during nuclear division in G. monilis, the arrangement of nuclei is much more disordered than the more even spacing before division, and there are no obvious microtubules to maintain the spacing between the nuclei. This suggests that in certain multinucleate algae, regular nuclear spacing is restored after one round of division by other factors that are not required during mitosis.
Most of the key processes that characterize living eukaryotic cells involve intracellular motility, and it is well established for both plant and animal cells that intracellular motility depends on cytoskeletal elements. In red algae, cytoplasmic streaming generally does not occur, and studies of cell division and the role of the cytoskeleton have mainly focused on microfilaments, which are relatively easy to stain (Garbary et al. 1992, McDonald et al. 1992, 1993, Kim and Kim 1999, Wilson et al. 2002, Kim et al. 2022), and studies of microtubules using antibodies and inhibitors have only recently become possible (Moon et al. 2022). In this study, we have developed a method for staining microtubules using fluorescent antibodies in G. monilis, and we expect that further studies of changes in cytoskeletal structure in response to Ca2+-mediated ROS signaling will allow us to better understand the role of the cytoskeleton as a mediator of cellular responses in red algae.
ACKNOWLEDGEMENTS
The authors express their sincere thanks to Prof. Joe Zuccarello for his valuable comments. This work was supported by the management of Marine Fishery Bio-resources Center (2023) funded by the National Marine Biodiversity Institute of Korea (MABIK) and by Development of technology for biomaterialization of marine fisheries by-products of Korea Institute of Marine Science & Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (KIMST-20220128) and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2019M3C1B7025093).
Notes
The authors declare that they have no potential conflicts of interest.