Five phototrophic Scrippsiella species lacking mixotrophic ability and the extended prey spectrum of Scrippsiella acuminata (Thoracosphaerales, Dinophyceae)
Article information
Abstract
Mixotrophic dinoflagellates act as primary producers, prey, and predators in marine planktonic food webs, whereas exclusively autotrophic dinoflagellates are primary producers and prey. Species of the dinoflagellate genus Scrippsiella are commonly found in marine ecosystems and sometimes cause harmful red tides. Among the 28 formally described Scrippsiella species, S. acuminata has been found to be mixotrophic and two unidentified species have been found to be mixotrophic. To determine whether the other species in this genus are similarly mixotrophic, the mixotrophic ability of S. donghaiensis SDGJ1703, S. lachrymosa SLBS1703, S. masanensis SSMS0908, S. plana SSSH1009A, and S. ramonii VGO1053 was explored using 15 potential prey items, including 2-μm fluorescently labeled microspheres (FLM) and heterotrophic bacteria (FLB), the cyanobacterium Synechococcus sp., and various microalgal prey species. The ability of S. acuminata to feed on FLM and FLB was also investigated. We found that S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, and S. ramonii did not feed on any potential prey tested in this study, indicating a lack of mixotrophy. However, S. acuminata fed on both FLM and FLB, confirming its mixotrophic ability. These results lowered the proportion of mixotrophic species relative to the total number of tested Scrippsiella species for mixotrophy from 100% to 29–38%. Owing to its mixotrophic ability, S. acuminata occupies an ecological niche that is distinct from that of S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, and S. ramonii.
INTRODUCTION
Mixotrophy, a combination of autotrophy and heterotrophy, is observed in many marine flagellates, dinoflagellates, and ciliates (Stoecker et al. 1997, Burkholder et al. 2008, Esteban et al. 2010, Jeong et al. 2010a, Lee et al. 2014a). Mixotrophs play diverse roles as primary producers, prey, and predators in marine ecosystems, whereas exclusively autotrophic organisms play the roles of primary producers and prey (Burkert et al. 2001, Jeong et al. 2012). Mixotrophy increases the growth rate of protists (Li et al. 2000, Smalley et al. 2003, Jeong et al. 2015, 2021, Ok et al. 2019, Kang et al. 2020, You et al. 2020), causes horizontal gene transfer, and is the main driving force in the evolution of photosynthetic organisms (Bhattacharya et al. 2004, Wisecaver et al. 2013, Hehenberger et al. 2019). Therefore, understanding mixotrophy is of ecological and evolutionary importance.
Dinoflagellates are ubiquitous protists in marine environments (Luo et al. 2021, Ok et al. 2021, Morquecho et al. 2022), and often form red tides or harmful algal blooms that can cause human diseases and massive mortality in shellfish, finfish, and mammals (Shumway 1990, Hallegraeff 1992, Flewelling et al. 2005, Jeong et al. 2017, Sakamoto et al. 2021). The trophic modes, growth and ingestion rates, and mortality rates due to predation of dinoflagellate species should be investigated to understand and predict the outbreak of dinoflagellate red tides (Franklin et al. 2006, Jeong et al. 2015). In the last three decades, many dinoflagellate species previously thought to be autotrophs were reclassified as mixotrophs (Bockstahler and Coats 1993, Stoecker et al. 1997), including many species that form red tides (Jeong et al. 2005a, 2005b, Burkholder et al. 2008, Park et al. 2013, Flynn et al. 2018). Approximately 90% of the dinoflagellates that form global red tides are mixotrophs (Jeong et al. 2021); however, less than 10% of the approximately 1,200 phototrophic dinoflagellates have been tested for mixotrophy (Stoecker et al. 1997, Jeong et al. 2005a, 2005b, Park and Kim 2010, Lee et al. 2014b, 2015, Lim et al. 2018, 2019). Therefore, understanding the ecological and genetic characteristics and red tide dynamics of a phototrophic dinoflagellate species requires an examination of their mixotrophic ability. Moreover, the prey species of mixotrophic dinoflagellates should be identified.
Since the description of the genus Scrippsiella by Balech (1959) with the type species S. sweeneyae, 28 species have been formally described (Hoppenrath et al. 2014, Guiry and Guiry 2023) (Supplementary Table S1). The species in the genus Scrippsiella have a global distribution; however, only S. acuminata (previously S. trochoidea) has caused red tides in the waters of many countries (Hallegraeff 1992, Pitcher et al. 2007, Pitcher and Joyce 2009, Soehner et al. 2012, Jeong et al. 2021). Red tides dominated by Scrippsiella spp. can cause fish mortality through hypoxia (Hallegraeff 1992); therefore, to minimize losses caused by Scrippsiella red tides, the ecophysiology and population dynamics of each Scrippsiella species should be understood (Jeong et al. 2015). Of the formally described and unidentified species in this genus, only S. acuminata and two unidentified Scrippsiella species have been confirmed to be mixotrophic (Jacobson and Anderson 1996, Jeong et al. 2005a, 2005b, Coats et al. 2020). Therefore, the mixotrophic abilities of other Scrippsiella species should be investigated. S. acuminata cells have been reported to feed on the cyanobacterium Synechococcus sp., prymnesiophyte Isochrysis galbana, cryptophytes Rhodomonas salina and an unidentified species, raphidophyte Heterosigma akashiwo, and phototrophic dinoflagellates Amphidinium carterae and Prorocentrum cordatum (Jeong et al. 2005a, 2005b). Furthermore, an unidentified Scrippsiella sp., which was collected from the waters off Korea, has been reported to feed on the tintinnid ciliate Helicostomella longa using a feeding tube (Coats et al. 2020). Another unidentified Scrippsiella sp. isolated from West Boothbay Harbor, Maine, United States was reported to possess food vacuoles (Jacobson and Anderson 1996). Heterotrophic bacteria are common in almost all marine ecosystems (Caron et al. 1982, Kjelleberg et al. 1987, Van Wambeke et al. 2000, Seong et al. 2006, Sanz-Sáez et al. 2020, Vijayan et al. 2022) and serve as prey for many mixotrophic dinoflagellates (Seong et al. 2006, Jeong et al. 2010a, 2012, Lee et al. 2014b, Millette et al. 2017). Thus, the ability of each Scrippsiella species to feed on heterotrophic bacteria warrants investigation.
In the present study, the mixotrophic abilities of five Scrippsiella species (Scrippsiella donghaiensis SDGJ1703, S. lachrymosa SLBS1703, S. masanensis SSMS0908, S. plana SSSH1009A, and S. ramonii VGO1053) were identified after they were provided 2-μm fluorescently labeled microspheres (FLM), fluorescently labeled heterotrophic bacteria (FLB), Synechococcus sp., and 12 microalgal species as potential prey. Whether S. acuminata STKP9909 can feed on FLM and FLB was also investigated. The present study provides a basis for understanding the ecological roles of these Scrippsiella species in marine ecosystems and their evolution within the genus Scrippsiella.
MATERIALS AND METHODS
Preparation of experimental organisms
Scrippsiella acuminata STKP9909, S. donghaiensis SDGJ1703, S. lachrymosa SLBS1703, and S. masanensis SSMS0908 were isolated from the waters off Kunpho in September 1999, Gijang in March 2017, Busan in March 2017, and Masan in August 2009, respectively. A clonal culture of each of the four species was established using two serial single-cell isolations (Kim et al. 2019, Lee et al. 2019) (Table 1). To isolate and culture S. plana SSSH1009A, surface sediment samples were collected from Shiwha Bay, Korea, in September 2010 using an Ekman Grab (Wildco; Wildlife Supply Company, Buffalo, NY, USA) (Table 1). The collected sediments were stored at 4°C in the dark and incubated as described by Jeong et al. (2014). A clonal culture of S. plana SSSH1009A was established using two serial single-cell isolations from the incubated sediment samples. A culture of S. ramonii VGO1053 that was originally collected from Ebro Delta in Spain was obtained from the Culture Collection of Microalgae (CCVIEO) of the Instituto Español de Oceanografía in Vigo (Table 1). All Scrippsiella cultures were transferred every two weeks in 250- or 800-mL flat culture flasks containing fresh F/2-Si medium (Guillard and Ryther 1962) and maintained in a 14 h : 10 h light : dark cycle under 50–100 μmol photons m−2 s−1 from a cool-white fluorescent light at 20°C.
The microalgal species selected as potential prey were maintained under the same light and temperature conditions as the Scrippsiella species (Table 2). The cyanobacterium Synechococcus sp. was maintained under 5–10 μmol photons m−2 s−1 from cool-white fluorescent light with a 14 h : 10 h light : dark cycle at 20°C.
Feeding occurrence test
Experiments 1–4 were designed to explore whether S. donghaiensis SDGJ1703, S. lachrymosa SLBS1703, S. masanensis SSMS0908, S. plana SSSH1009A, and S. ramonii VGO1053 can feed on the FLM (Expt 1: diameter = 2 μm; 18604 Fluoresbrite YG Microspheres; Polysciences Inc., Warrington, PA, USA), FLB (Expt 2), Synechococcus sp. (Expt 3), or microalgal prey species (Expt 4). Whether S. acuminata STKP9909 can feed on the FLM and FLB was also investigated.
In Expt 1, approximately 3 × 107 FLM were added to a 30-mL polycarbonate (PC) rounded bottle containing either S. acuminata, S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, or S. ramonii. One experimental bottle (one Scrippsiella species + FLM), one prey control bottle (FLM only), and one predator control bottle (one Scrippsiella species only) were included in each experimental run. The bottles were incubated on a plankton wheel rotating at 0.9 rpm (0.00017 ×g) at 20°C under a 14 h : 10 h light-dark cycle (20 μmol photons m−2 s−1). After 2 and 24 h of incubation, 3-mL aliquots were removed from each bottle and transferred to confocal dishes (SPL100350; SPL Life Sciences Co., Ltd., Pocheon, Korea). To observe Scrippsiella spp. feeding on the FLM, the protoplasm of 200 cells of each target Scrippsiella species was carefully observed under an inverted microscope (Zeiss Axiovert 200M; Carl Zeiss Ltd., Göttingen, Germany) and photographs were taken using a digital camera (Zeiss Axiocam 506; Carl Zeiss Ltd.) attached to the microscope at 1,000× magnification.
To prepare Expt 2, marine heterotrophic bacterial cells were obtained by filtering non-axenic cultures of S. acuminata, S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, and S. ramonii. Aliquots of 300–2,000 mL from each Scrippsiella culture were serially filtered through 5.0-μm and 1.2-μm pore-sized filter papers (Merck Millipore, Burlington, MA, USA). The filtrates containing only bacterial cells were centrifuged at 3,000 rpm (2,063 ×g) for 30 min at 4°C (Labogene 1696R; Gyrozen Co., Gimpo, Korea) in Falcon tubes (Falcon; Corning, New York, NY, USA). The centrifuged bacterial cells were fluorescently labeled with 5-(4,6-dichlorotriazin-2-yl) amino fluorescein hydrochloride (D0531; Sigma-Aldrich, St. Louis, MO, USA) according to the method of Sherr et al. (1987) and then stored in the dark at 4°C until use. The FLB cells were resuspended in the culture medium using a sonicator (Bransonic cleaner 5510E-DTH; Branson Ultrasonics, Danbury, CT, USA) for 10 min and then filtered through a 3-μm pore-sized filter paper (Merck Millipore) to remove aggregated bacterial cells. Each Scrippsiella spp. was tested using FLB collected from its own culture.
In Expts 2 and 3, approximately 3 × 107 FLB (Expt 2) or Synechococcus cells (Expt 3) were added to 30-mL PC rounded bottles containing each Scrippsiella species (Table 3). After 1 and 4 h of incubation, 3-mL aliquots were removed from each bottle, transferred into confocal dishes (SPL Life Sciences Co., Ltd.), and carefully observed as described above.
In Expt 4, dense cultures of S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, and S. ramonii were added to the wells of 6-well plates containing each algal prey species. One experimental well (one Scrippsiella species + one microalgal prey species), one prey control well (microalgal prey species only), and one predator control well (one Scrippsiella species only) were established, and the plate was cultured in a 14 h : 10 h light : dark cycle (20 μmol photons m−2 s−1 ) at 20°C. After 2 and 24 h of incubation, 3-mL aliquots were removed from each experimental well and transferred to confocal dishes (SPL Life Sciences Co., Ltd.). The protoplasm of 200 cells of target Scrippsiella spp. was observed using an inverted microscope (Carl Zeiss Ltd.) and photographed at 1,000× magnification.
DNA sequencing and phylogenetic analysis
The large subunit (LSU) rDNA sequences of S. acuminata STKP9909, S. donghaiensis SDGJ1703, and S. plana SSSH1009A were not previously reported and that of S. ramonii VGO1053 was previously partially reported. To obtain these sequences, 1 μL from each culture was removed and added into 0.2-mL polymerase chain reaction (PCR) tubes with 38.75 μL deionized sterile distilled water. Subsequently, a mixture of 1 μL dNTP mix, 0.25 μL F-StarTaq DNA polymerase, 5 μL 10× F-StarTaq buffer (BioFACT Co., Ltd., Daejeon, Korea), and 4 μL of each forward-reverse primer set needed for amplification of the LSU rDNA region (Set 1, ITSF2 and LSU500R; Set 2, D1R and 1483R) was added to the test tubes. The sequences of the primers used are as follows: ITSF2, 5′-TAC GTC CCT GCC CTT TGT AC-3′; D1R, 5′-ACC CGC TGA ATT TAA GCA TA-3′; LSU500R, 5′-CCC TCA TGG TAC TTG TTT GC-3′; 1483R, 5′-GCT ACT ACC ACC AAG ATC TGC-3′ (Scholin et al. 1994, Daugbjerg et al. 2000, Litaker et al. 2003). PCR was conducted using an AllInOne Cycler (Bioneer, Daejeon, Korea) under the same thermal conditions as described by You et al. (2023). The PCR products were purified using an AccuPrep DNA Purification Kit (Bioneer) and then sequenced using an ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Sequences were aligned and manually edited using ContigExpress (Infomax, Frederick, MD, USA).
For phylogenetic analysis, LSU rDNA sequences from 19 Scrippsiella spp. were obtained from this study and GenBank and aligned using MEGA v4 (Tamura et al. 2007). A LSU rDNA phylogenetic tree was constructed using Bayesian (the default GTR + G + I model in MrBayes v3.1) (Ronquist and Huelsenbeck 2003) and maximum likelihood analyses (the default GTRGAMMA model in RAxML 7.0.3 program) (Stamatakis 2006). The assumed empirical nucleotide frequencies of the LSU rDNA comprised a substitution rate matrix where A–C = 0.0344, A–G = 0.1778, A–T = 0.0562, C–G = 0.0163, C–T = 0.6361, and G–T = 0.0792. The rates were assumed to follow a gamma distribution with a shape parameter of 0.4747 for the variable sites. The proportion of the sites that were assumed to be invariable was 0.2041.
RESULTS
Feeding occurrence of the Scrippsiella spp
In Expt 1, the 2-μm FLM were found in the protoplasm of S. acuminata STKP9909 cells, although they were rarely observed (Table 3, Fig. 1). However, no FLM were observed in the protoplasm of observed S. donghaiensis SDGJ1703, S. lachrymosa SLBS1703, S. masanensis SSMS0908, S. plana SSSH1009A, or S. ramonii VGO1053 cells (Table 3, Fig. 2). Similarly, in Expt 2, FLBs were found in the protoplasm of S. acuminata STKP9909 cells, although they were rarely observed (Table 3, Fig. 3A & B); however, S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, and S. ramonii did not feed on FLB (Table 3, Fig. 3C–L). Moreover, in Expt 3, S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, and S. ramonii did not feed on Synechococcus sp. (Table 3).
Scrippsiella acuminata STKP9909 (Sa) not fed (A & B) or fed (C–F) fluorescently labeled microspheres (FLM). Micrographs A, C, and E were taken under a light microscope and B, D, and F under an epifluorescence microscope. Green arrows indicate Sa cells, black and white arrows indicate not ingested FLM, and red arrows indicate ingested FLM within Sa cells. Scale bars represent: A–F, 10 μm.

Scrippsiella donghaiensis SDGJ1703 (Sd; A & B), S. lachrymosa SLBS1703 (Sl; C & D), S. masanensis SSMS0908 (Sm; E & F), S. plana SSSH1009A (Sp; G & H), and S. ramonii VGO1053 (Sr; I & J), not fed fluorescently labeled microspheres (FLM). Micrographs A, C, E, G, and I were taken under a light microscope and those in B, D, F, H, and J under an epifluorescence microscope. Green arrows indicate Scrippsiella cells; black and white arrows indicate not ingested FLM. Scale bars represent: A–J, 10 μm.
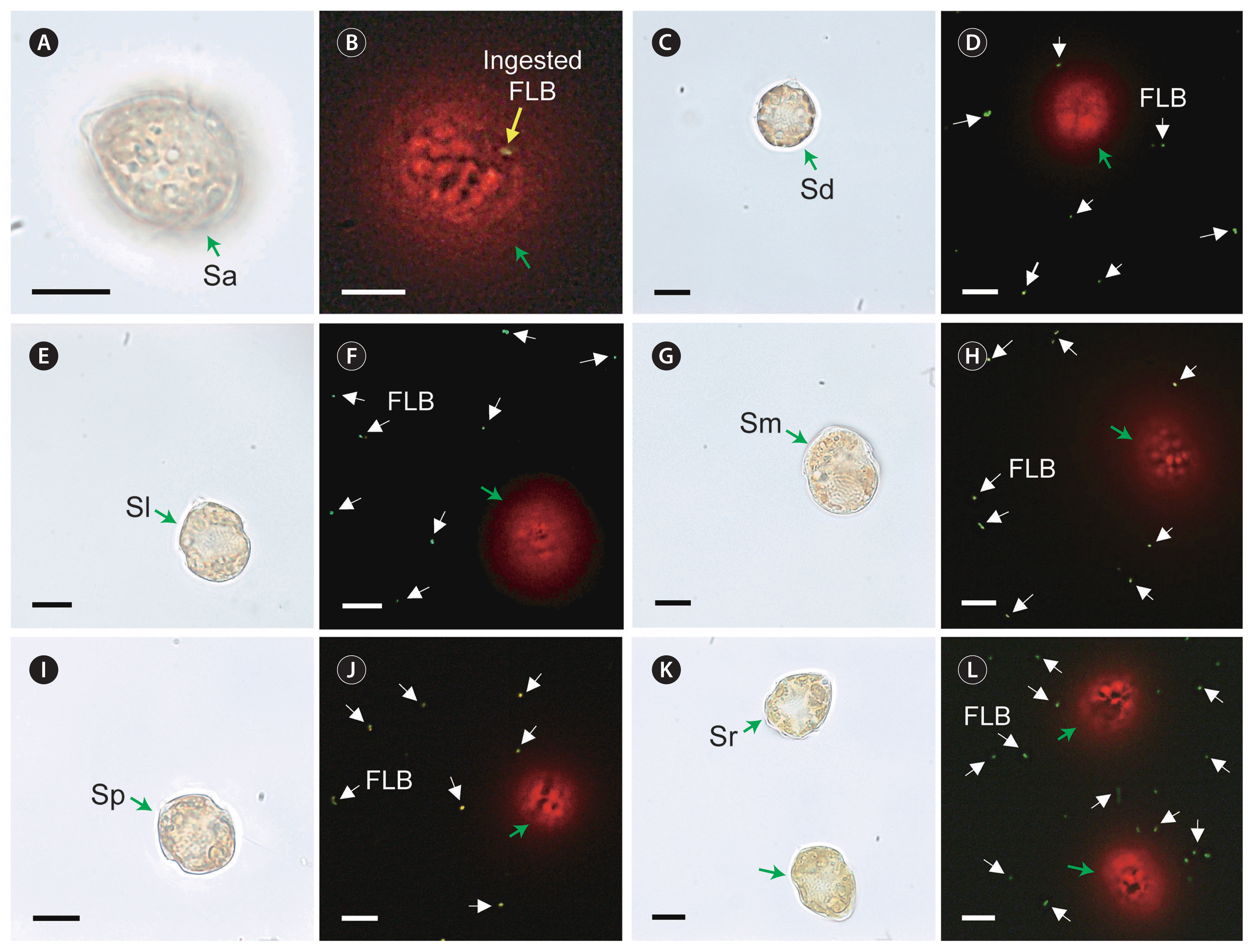
A Scrippsiella acuminata STKP9909 (Sa; A & B) cell feeding on the fluorescently labeled heterotrophic bacteria (FLB) and S. donghaiensis SDGJ1703 (Sd; C & D), S. lachrymosa SLBS1703 (Sl; E & F), S. masanensis SSMS0908 (Sm; G & H), S. plana SSSH1009A (Sp; I & J), and S. ramonii VGO1053 (Sr; K & L) not feeding on the FLB. Micrographs A, C, E, G, I, and K were taken under a light microscope and B, D, F, H, J, and L under an epifluorescence microscope. Green arrows, Scrippsiella cells; a yellow arrow, ingested FLB within a Scrippsiella cell; white arrows, not ingested FLB. Scale bars represent: A–L, 10 μm.
In Expt 4, S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, and S. ramonii did not feed on any of the microalgal prey species, which included the prymnesiophyte Isochrysis galbana, prasinophyte Pyramimonas sp. (PSSH1204), cryptophytes Teleaulax amphioxeia (TSGS0202), Storeatula major (SSSH1103), and Rhodomonas salina, raphidophyte Heterosigma akashiwo (HAKS9905), and phototrophic dinoflagellates Heterocapsa rotundata (HRSH1201), Heterocapsa minima (HMMJ1604), Amphidinium carterae (SIO PY-1), Prorocentrum cordatum (PMKS9906), Prorocentrum donghaiense (PDYS1407), Prorocentrum micans (PMSH0910), and Akashiwo sanguinea (ASUSA) (Table 3, Fig. 4). Furthermore, S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, and S. ramonii did not show any attack behaviors toward the prey items, inhibit their swimming, or lyse the prey.

Cells of Scrippsiella donghaiensis SDGJ1703 (Sd; A & B), S. lachrymosa SLBS1703 (Sl; C & D), S. masanensis SSMS0908 (Sm; E & F), S. plana SSSH1009A (Sp; G & H), and S. ramonii VGO1053 (Sr; I & J), not feeding on the dinoflagellate Amphidinium carterae (Ac). A, C, E, G, and I, Scrippsiella cells without Ac (control). B, D, F, H, and J, Scrippsiella cells with Ac (expt). Green arrows indicate Scrippsiella cells and black arrows indicate Ac cells. Scale bars represent: A–J, 10 μm.
Phylogenetic analysis of Scrippsiella spp
We obtained four novel LSU rDNA sequences from S. acuminata STKP9909, S. donghaiensis SDGJ1703, S. plana SSSH1009A, and S. ramonii VGO1053 and deposited them in GenBank under the accession numbers OQ266790, OQ266882, OQ266885, and OQ275008 for S. acuminata STKP9909, S. donghaiensis SDGJ1703, S. plana SSSH1009A, and S. ramonii VGO1053, respectively. A phylogenetic tree generated using LSU rDNA showed that S. acuminata, S. lachrymosa, and S. ramonii belong to a large clade along with S. spinifera, S. kirschiae, S. trifida, S. bicarinata, S. erinacea, S. sweeneyae, Scrippsiella sp. JKG47-2, S. precaria, and S. irregularis (Fig. 5). This clade included not only the two mixotrophic species, S. acuminata STKP9909 and Scrippsiella sp. JKG47-2, but also S. lachrymosa and S. ramonii, which lack mixotrophic abilities. The clades that included S. donghaiensis SDGJ1703, S. masanensis SSMS0908, and S. plana SSSH1009A, which lack mixotrophic abilities, were divergent in the phylogenetic tree.
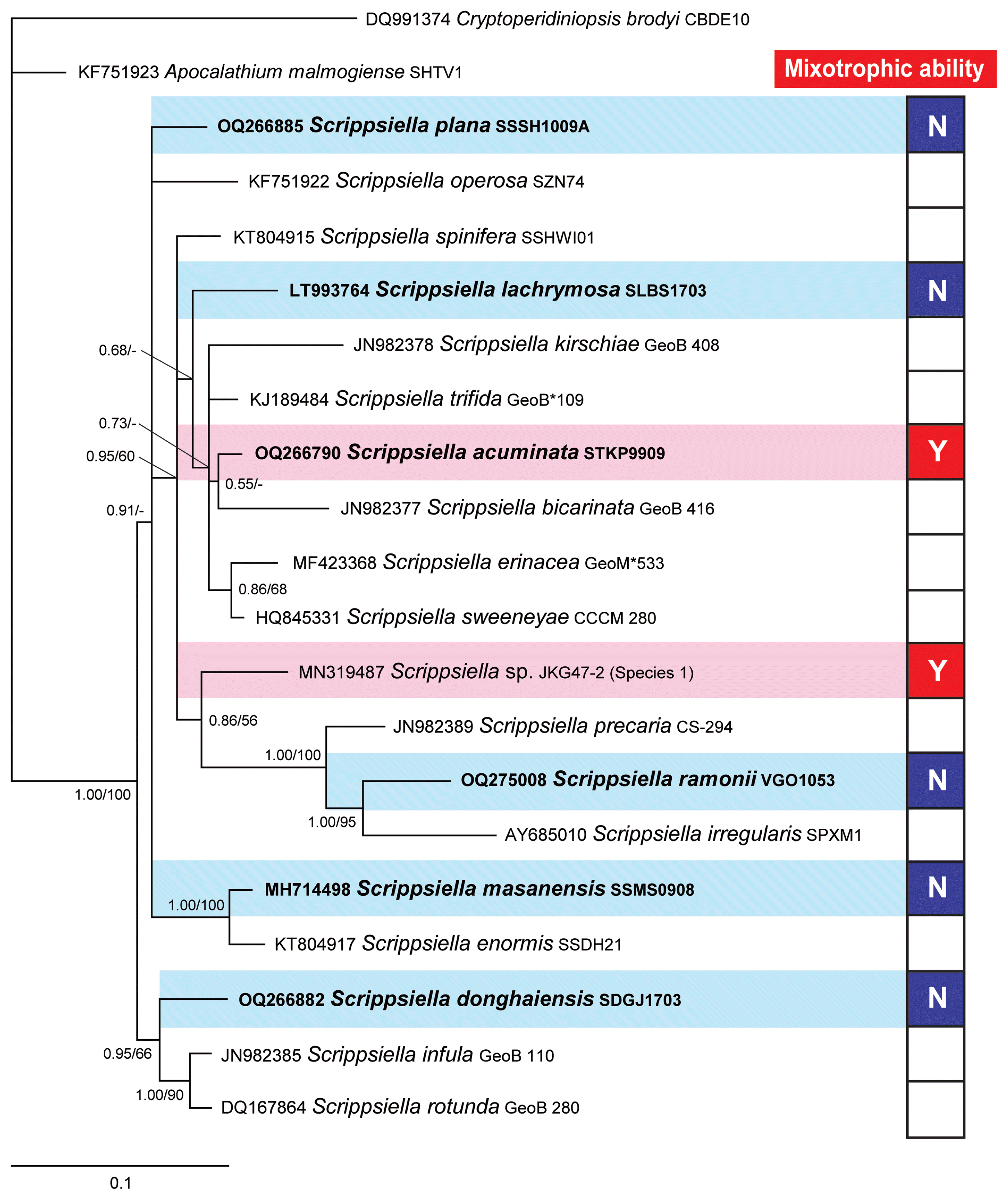
Consensus Bayesian tree based on 601-bp aligned positions of the large subunit regions from 19 species within the genus Scrippsiella. Sequences from Cryptoperidiniopsis brodyi and Apocalathium malmogiense were used as an outgroup. The number of character changes are proportional to branch lengths and indicate the maximum likelihood bootstrap values (right) and Bayesian posterior probability (left); posterior probabilities ≥0.5 are shown; the species names are followed by the strain names of each species. The species tested in this study are shown in bold. Red boxes (Y) indicate mixotrophic Scrippsiella species; blue boxes (N) indicate Scrippsiella species without mixotrophic ability; and white boxes represent Scrippsiella species that were not tested for mixotrophy. Data of the presence or absence of mixotrophic ability for species within the genus Scrippsiella were obtained from this study, Jeong et al. (2005a, 2005b), and Coats et al. (2020).
DISCUSSION
The present study reports, for the first time, that S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, and S. ramonii lack mixotrophic abilities. Previous studies reported that three Scrippsiella species, S. acuminata and two unidentified Scrippsiella spp. were mixotrophic (Jacobson and Anderson 1996, Jeong et al. 2005a, 2005b, Coats et al. 2020). The results of the present study lower a proportion of mixotrophic species relative to the total Scrippsiella species tested for mixotrophy. In the phylogenetic tree based on LSU rDNA, the unidentified Scrippsiella species isolated from Korean waters was divergent from S. acuminata, indicating a distinct species. If the two unidentified Scrippsiella species from the United States and Korean waters are distinct from each other and from S. acuminata, the proportion of mixotrophic species relative to that of the total Scrippsiella species tested for mixotrophy is 38% (3 of 8 tested species). However, if the unidentified Scrippsiella species from the United States is S. acuminata or the unidentified Scrippsiella species from Korean waters, the proportion of the number of mixotrophic species relative to that of the total Scrippsiella species tested for mixotrophy is 29% (2 of 7 tested species). The mixotrophic abilities of species in the dinoflagellate genera Alexandrium, Paragymnodinium, and Karenia have also been reported (Lim et al. 2019, Yokouchi et al. 2022, Ok et al. 2023) (Table 4). The proportion of mixotrophic species relative to the total species tested for mixotrophy was 44% (7 of 16 tested species) in the genus Alexandrium, 60% (3 of 5 tested species) in the genus Paragymnodinium, and 40% (2 of 5 tested species) in the genus Karenia (Table 4). Thus, the proportion of mixotrophic species in the genus Scrippsiella was slightly lower than that in the genera Karenia or Alexandrium and considerably lower than that in the genus Paragymnodinium. Among the formally described 34, 10, and 28 species in the genera Alexandrium, Karenia, and Scrippsiella, respectively, the mixotrophic abilities of 16, 5, and 6 species have been explored. Thus, the proportion of mixotrophic species relative to the total species tested for mixotrophy may be changed.

Number of species having or lacking the mixotrophic ability in the dinoflagellate genera Scrippsiella, Alexandrium, Karenia, and Paragymnodinium
In the phylogenetic tree based on the LSU rDNA sequences of 19 Scrippsiella species, a large clade included S. acuminata and Scrippsiella sp. JKG47-2, which have mixotrophic abilities, and S. lachrymosa and S. ramonii, which lack mixotrophic abilities. However, three clades, S. donghaiensis SDGJ1703, S. masanensis SSMS0908, and S. plana SSSH1009A, that lack mixotrophic abilities diverged from the large clade. Thus, the ancestral species of Scrippsiella may have lacked feeding ability and acquired it later through evolution. However, to confirm this hypothesis, the mixotrophic abilities of other Scrippsiella spp. need to be examined.
S. acuminata was able to feed on the cyanobacterium Synechococcus sp., a prymnesiophyte, cryptophytes, raphidophytes, and phototrophic dinoflagellates that were <12.1 μm in equivalent spherical diameter (Jeong et al. 2005a, 2005b). The results of the present study expand the range of prey items of S. acuminata to include FLB. Heterotrophic bacteria are ubiquitous (Caron et al. 1982, Seong et al. 2006, Sanz-Sáez et al. 2020, Vijayan et al. 2022); thus, the ability of S. acuminata to feed on heterotrophic bacteria may be critical for the survival of this dinoflagellate species under conditions of inorganic nutrient depletion. Heterotrophic bacteria usually have high phosphorus : nitrogen ratios (Vadstein et al. 1988, Tezuka 1990) and some cyanobacteria can conduct nitrogen fixation (Mitsui et al. 1987, Zehr 2011). Therefore, S. acuminata may obtain phosphorus and nitrogen for their growth by feeding on heterotrophic bacteria and cyanobacteria in offshore or oceanic waters (Jeong et al. 2010b).
S. acuminata has a global distribution and can cause red tides in many countries (Moncheva et al. 2001, Pitcher et al. 2007, Gárate-Lizárraga et al. 2009, Jeong et al. 2021, Tsikoti and Genitsaris 2021). However, S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, or S. ramonii, which lack mixotrophic abilities, has caused few or no red tides (Jang et al. 2022). Thus, the mixotrophic ability of S. acuminata may allow it to cause red tides in several marine ecosystems.
S. acuminata is preyed upon by the common heterotrophic dinoflagellates Oxyrrhis marina, Gyrodinium dominans, Polykrikos kofoidii, Oblea rotunda, and Pfiesteria piscicida, ciliates Tiarina fusus and Strombidinopsis sp., copepods Acartia omorii, Calanus helgolandicus, Calanus pacificus, and Temora longicornis, and larvae of the mussel Mytilus galloprovincialis (Gill and Harris 1987, Hassett and Landry 1990, Jeong et al. 2002, 2004, Shin et al. 2003, Kim et al. 2019). Thus, S. acuminata plays an ecological role as a primary producer and predator of heterotrophic bacteria, cyanobacteria, and diverse microalgae, and serves as a suitable prey item for many heterotrophic protists in marine ecosystems. S. donghaiensis, S. lachrymosa, and S. masanensis are also consumed by O. marina, G. dominans, P. kofoidii, O. rotunda, P. piscicida, and Strombidinopsis sp. (Kim et al. 2019). Thus, S. donghaiensis, S. lachrymosa, and S. masanensis may play ecological roles as primary producers and prey for heterotrophic protists in marine ecosystems, and owing to its mixotrophic ability, S. acuminata has a different ecological niche from that of S. donghaiensis, S. lachrymosa, S. masanensis, S. plana, and S. ramonii.
ACKNOWLEDGEMENTS
We thank editors and reviewers for their valuable comments. This research was supported by Korea Institute of Marine Science & Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (20230018) and the National Research Foundation of Korea funded by the Ministry of Science and ICT (NRF-2021M3I6A1091272; NRF-2021R1A2C1093379) award to HJJ.
Abbreviations
FLB
fluorescently labeled heterotrophic bacteria
FLM
fluorescently labeled microspheres
LSU
large subunit
rDNA
ribosomal DNA
Notes
The authors declare that they have no potential conflicts of interest.
SUPPLEMENTARY MATERIALS
List of the 28 formally described species of the genus Scrippsiella (https://www.e-algae.org).


