Based on morphology and molecular data, Palisada rigida comb. nov. and Laurencia decussata comb. et stat. nov. (Rhodophyta, Rhodomelaceae) are proposed
Article information
Abstract
Inspecting herbaria collections of Laurencia rigida highlighted frequent misidentifications between L. rigida and L. heteroclada f. decussata, two poorly studied taxa from Australia. Recent collections of DNA material, including from topotype material, allowed for re-examination of these two taxa using molecular techniques. Detailed morphological and molecular analyses based on two markers (rbcL and COI-5P) strongly supported these two taxa as being distinct from each other and requiring nomenclatural changes. Comprehensive morphological analyses highlighted features useful for accurate identifications. Interestingly, L. rigida was found to belong to the genus Palisada with evidence from both the morphology and molecular data. Therefore, this study proposed recognizing L. rigida as Palisada rigida comb. nov. Molecular data for L. heteroclada f. decussata on the other hand supported its separation from L. heteroclada, with too great a molecular distance to be considered a variety. Morphological characters that best separated P. rigida from L. decussata included seven characters; number of pericentral cells per vegetative axial segment, the presence of secondary pit connections, the presence of lenticular thickenings, tetrasporangia alignment, the presence of corps en cerise, holdfast morphology, and overall plant shape. Morphologically, L. heteroclada f. decussata was also separated from L. heteroclada, particularly by the following characteristics; ultimate branchlets morphologies, lower order branch lengths, primary axis and holdfast morphologies. Therefore, it was proposed that L. heteroclada f. decussata is recognized at a species level as L. decussata comb. et stat. nov.
INTRODUCTION
Laurencia complex species are common, intertidal marine red algae found throughout most of the world’s coastlines (Guiry and Guiry 2021). They are recognized by having depressed apical pits containing a single apical cell and a polysiphonous construction, which is only observable at branch apices due to extensive cortication. The Laurencia complex is currently comprised of eight genera; Osmundea Stackhouse (1809), Laurencia J. V. Lamouroux (1813), Corynecladia J. Agardh (1876), Chondrophycus (Tokida and Saito) Garbary and J. T. Harper (Garbary and Harper 1998), Palisada (Yamada) K. W. Nam (Nam 2007), Yuzurua (K. W. Nam) Martin-Lescanne (Martin-Lescanne et al. 2010), Laurenciella Cassano, Gil-Rodríguez, Sentíes, Díaz-Larrea, M. C. Oliveira and M. T. Fujii (Cassano et al. 2012), and Ohelopapa F. Rousseau, Martin-Lescanne, Payri and L. LeGall (Rousseau et al. 2017).
Laurencia was first described by Lamouroux in 1813 and almost immediately was reorganized to include new sections and subgenera (Stackhouse 1816). Osmundea was originally proposed in 1809 (Stackhouse 1809) but only recognized as a genus much later (Furnari and Serio 1993, Nam et al. 1994). Soon after, through the support of morphological cladistic analyses, the Laurencia subgenus Chondrophycus was raised to generic rank (Garbary and Harper 1998). The genus Palisada was not formally recognized until many years later. Instead Palisadae was introduced as a section within Laurencia (Saito and Womersley 1974). Palisadae was then raised to subgeneric rank (Nam 1999), and then subsequently to generic rank (Nam 2007) along with an introduction of new diagnostic morphological characters (Nam 2006). These characters were necessary to separate Palisada from the already recognized Chondrophycus genus (Garbary and Harper 1998, Nam 2007). Once Palisada was recognized there were four accepted genera within the Laurencia complex; Laurencia, Osmundea, Chondrophycus, and Palisada. All were established based on morphological features. The first two genera within the complex to be determined by molecular evidence (rbcL) were Yuzurua (Martin-Lescanne et al. 2010) and Laurenciella (Cassano et al. 2012). This early molecular research supported the continued recognition of the original four genera as well. In 2015, the genus Coronaphycus was established using both molecular (rbcL) and morphological evidence (Metti et al. 2015), but was later determined to be conspecific with Corynecladia J. Agardh (1876), which had priority (Cassano et al. 2019). The final genus to be established, as of this writing, using both molecular (rbcL and 5′ end of cytochrome c oxidase I large subunit [COI-5P]) and morphological data was the monotypic Ohelopapa (Rousseau et al. 2017). The genus Laurencia still contains the largest number of taxa, with 137 currently recognized species (Guiry and Guiry 2021).
Laurencia rigida J. Agardh (1876) is primarily known from Eastern Australia, but has been reported from the Indian Ocean, New Zealand, southern Asia and the Pacific Islands (Cribb 1958, Millar and Kraft 1993, Silva et al. 1996, Guiry and Guiry 2021). The type was described by J. Agardh with a location simply of ‘in oceano indico calidiore.’ It has been generally accepted that this refers to the warmer part of Australia, most likely somewhere along the Queensland coast (Yamada 1931, Saito and Womersley 1974, Millar 1990). In Australia, L. rigida has since been recorded from Queensland (Cribb 1958, Lewis 1984) and New South Wales (NSW) (Millar 1990, Millar and Kraft 1993, Zuccarello and West 2006). Laurencia rigida can be identified in part by its branching structure, including basal branches which are longer than branches found more apically along the primary axes, and having short ultimate branches, particularly in comparison to the long primary and secondary axes. However, a close inspection of L. rigida specimens lodged in Australian herbaria revealed that many specimens determined as L. rigida, particularly in NSW, more closely fit the current descriptions and type of Laurencia heteroclada f. decussata A. B. Cribb (1958). It also seemed that L. rigida specimens matching the type specimen actually may have better fit in the Palisada genus.
Laurencia heteroclada Harvey (1855) was originally described from Rottnest Island, Western Australia (WA). Since then, in Australia it has been recorded from; Queensland (Cribb 1958), Lord Howe Island (Millar and Kraft 1993 as L. filiformis f. heteroclada), WA, South Australia (SA), Victoria and Tasmania (Womersley 2003 as L. filiformis f. heteroclada). Laurencia heteroclada f. decussata was described by A. B. Cribb from Miami, south Queensland (Cribb 1958). He placed his Miami samples as a subset of L. heteroclada because of their similar stoloniferous holdfasts, rocky habitats in the lower littoral to upper sublittoral zones, overall sizes, and textures; both are fleshy and cartilaginous but not rigid. However, he described his taxa as a variety because of differences in distributions; L. heteroclada ranges from Redcliffe northwards, and L. heteroclada f. decussata from Miami southwards (Cribb 1958). Cribb also separated the two taxa due to differences in branching structures. Laurencia heteroclada f. decussata showed ultimate branchlets pressed up to the supporting branch and narrow column-like branching, whereas L. heteroclada did not.
In this study, fresh collections of L. heteroclada f. decussata and L. rigida from Australia allowed for both species to be re-examined in light of new molecular information to more accurately distinguish between them.
MATERIALS AND METHODS
Collections
The following taxa were collected from near their type localities; L. heteroclada f. decussata, L. rigida, and L. heteroclada. They were also collected from various other locations in Australia. Samples were dried in silica powder for molecular work and preserved in 4% formalin in seawater. Vouchers were pressed as well. Morphological studies were carried out on liquid preserved material and pressed vouchers. Liquid preserved material was stained with 1% aniline blue and 1% acetic acid solution, sectioned by hand, stained again, then fixed with a 50% karo solution. Microscopic observations were then made using a Zeiss compound microscope (Zeiss, Oberkochen, Germany). A BBT Krauss dissector microscope (BBT Krauss, Paris, France) was used to observe surface features of liquid preserved and pressed materials. Photos were taken with a Nikon coolpix4500 digital camera (Nikon, Tokyo, Japan) and microscope adapter lenses were used for slide photos. All slides, vouchers, silica dried material and liquid preserved material are stored at the National Herbarium of New South Wales (NSW). The type specimens of L. heteroclada f. decussata (BRI), L. rigida (LUND, MEL), and L. heteroclada (TCD) were examined, as well as other borrowed herbarium specimens.
DNA sequencing
The DNEasy Plant Mini Kit (Qiagen, Valencia, CA, USA) was used to extract genomic DNA from silica preserved samples. The JetQuick PCR Purification Kit (Genomed Co., Lohne, Germany) was used to purify DNA before and after amplification by polymerase chain reaction (PCR). The large subunit of RuBisCO (rbcL) gene region was amplified in one part using the primer pairs FrbcL_start_sh and RrbcS_st (Metti et al. 2013), or in two parts using FrbcL_start_sh and R_749 combined with F_749 and RrbcS_st (Metti 2017). The COI-5P gene region was amplified in one part using the primers GAZF1 and GAZR1 (Saunders 2005). Amplification methods followed those outlined in Metti et al. (2015) using a Corbett Palm thermocycler (Corbett Research, Mortlake, Australia). Amplified products were purified using the JetQuick PCR Purification Kit (Genomed Co.). Four microliters of purified PCR product was run on an agarose gel to visualize DNA concentrations. Primers used for the sequencing reactions were the same as used for amplification, for both gene markers. For the sequencing reactions, the method outlined in Metti et al. (2015) was followed using a Corbett Palm thermocycler (Corbett Research). The samples were precipitated and dried following the University of New South Wales (UNSW) Ramaciotti Centre ethanol/EDTA precipitation protocol. They were then sent to the UNSW in Sydney, Australia for sequencing. The Ramaciotti Centre sequencing protocol was followed for the 3730 Capillary Sequencer (Applied Biosystems, Scoresby, VIC, Australia).
Phylogenetic analysis
Raw sequence data was cleaned using Geneious 10.2.5 (https://www.geneious.com). Sequences were then aligned in BIOEDIT v7.2.0 for PC (Hall 1999) using the accessory application ClustalW Multiple Alignment program (Thompson et al. 1994), then visually checked and corrected. Additional relevant sequences were downloaded from GenBank (Clark et al. 2016), including outgroup sequences. Both the 5′ and 3′ ends were trimmed due to variable sequence lengths, resulting in a final length of 1,284 bp for the rbcL alignment, and 568 bp for the COI-5P alignment. A total of 76 rbcL sequences, and 62 COI-5P sequences were included in each alignment. Newly generated sequences (21 for rbcL and 19 for COI-5P) were deposited in GenBank (http://www.ncbi.nlm.nih.gov). All sequence and collection data were reported in Supplementary Table S1.
Pairwise distances (p.d.) were calculated using uncorrected ‘p’ distances in PAUP for PC (v.4.0a 169) (Swofford 2017). The online CIPRES Gateway (Miller et al. 2010) was used to run the maximum likelihood (ML) and Bayesian analyses (BI) for rbcL. RaxML (v8.2.12) (Stamatakis 2014) was used for inferring ML phylogenies as well as estimating node support with 1,000 bootstrap replicates. The General Time Reversible (GTR) evolutionary substitution model was set by RaxML, using a gamma-distributed rate variation. For the rbcL gene the assumed nucleotide frequencies were freq(A) = 0.315503, freq(C) = 0.157755, freq(G) = 0.211786, and freq(T) = 0.314956. The substitution rate matrix was A-C = 1.846410, A-G = 7.736281, A-T = 2.675699, C-G = 1.452319, C-T = 17.388636, and G-T = 1.000000. For rbcL MrBayes 3.2.7a (Ronquist et al. 2012) was used for the BI analyses using the standard GTR evolutionary model. One hot and three cold chains of the Markov chain Monte Carlo were used. The analyses started with a random tree, and ran for 1,000,000 runs, with sampling every 1,000 generations. The first 25% of generated trees were discarded as the burn-in. A 50% majority rule consensus tree was calculated from the remaining trees and posterior probabilities determined. The COI-5P sequences were analyzed using neighbour-joining (NJ) and maximum parsimony (MP) methods. Both were performed using the software PAUP for PC (v.4.0 beta10) (Swofford 2003). Node support was estimated using 2,000 bootstrap replicates for the NJ analysis and 1,000 for the MP analysis.
RESULTS
Molecular
Both rbcL and COI-5P trees were congruent in showing L. heteroclada f. decussata and L. rigida as supported clades within the Laurencia complex.
rbcL
The rbcL tree (Fig. 1) showed the following generic clades to be fully supported; Laurencia, Yuzurua, Laurenciella, Palisada, Osmundea, and Chondrophycus. The Corynecladia clade was highly to moderately supported (ML bootstrap = 92%, BI posterior probability = 0.97), and the Ohelopapa clade was unsupported with only one sequence. Both the Laurencia sensu stricto (s.s.) clade and the Palisada clade were fully supported within the complex.

The RaxML maximum likelihood (ML) tree inferred from rbcL sequence data. Bayesian posterior probabilities and ML bootstrap values (1,000 replicates) greater than 0.70/70 are shown. Bold and * indicates full support. Sequences resulting from this study are in bold text, and those of topotype material are indicated with ^.
The L. rigida clade was comprised of 10 sequences. This included a topotype sequence from Queensland (GenBank No. OM328131) that morphologically matched well with the type specimens. The L. rigida clade resulted in a fully supported, monophyletic clade within Palisada. Intraspecific p.d. within this clade were 0.00–0.55%. This was well within species limits generally accepted for rbcL which are usually <2.00% (Nam et al. 2000, Díaz-Larrea et al. 2007, Cassano et al. 2009, Metti et al. 2015). The closest sequence to the L. rigida clade was the topotype sequence of Palisada perforata (GenBank No. EU256325.1 as Palisada papillosa) from the Canary Islands. This relationship was unsupported and the pairwise distance between the topotype sequence of L. rigida and this P. perforata sequence was 2.80%. This was only slightly higher than accepted species limits, but much larger than p.d. within the L. rigida clade itself (≤0.55%).
The L. heteroclada f. decussata clade contained 11 sequences, and nested within the Laurencia s.s. clade. This clade included a topotype sequence from Queensland (GenBank No. OM328126) that matched very well with the type specimens. The intraspecific p.d. for the L. heteroclada f. decussata clade ranged from 0.00–0.32%. Results showed L. heteroclada f. decussata separated from the L. heteroclada topotype sequence, with a pairwise distance of 3.82%.
The L. heteroclada f. decussata clade was also separated from the L. rigida clade. The two topotype sequences were separated by a p.d. of 9.58%, supporting their separation at a generic level, which is generally accepted to be above 8.00% (Cassano et al. 2012, Metti et al. 2015). The closest sequences to L. heteroclada f. decussata was the clade containing L. yamadana, L. caduciramulosa, L. filiformis, and L. natalensis. The relationship between this clade and L. heteroclada f. decussata clade was strongly supported (ML bootstrap = 99%, BI posterior probability = 1.00). The closest pairwise distance between this clade and L. heteroclada f. decussata was with L. natalensis (p.d. = 2.32%). This is only slightly higher than accepted species limits, but again much larger than p.d. within the L. heteroclada f. decussata clade itself (≤0.32%).
COI-5P
The COI-5P results were congruent with the rbcL results. However, most generic clades were unsupported. Only the Chondrophycus and Laurenciella clades were moderately supported for NJ (Fig. 2). However, when looking at the L. heteroclada f. decussata and L. rigida clades, both were highly supported.
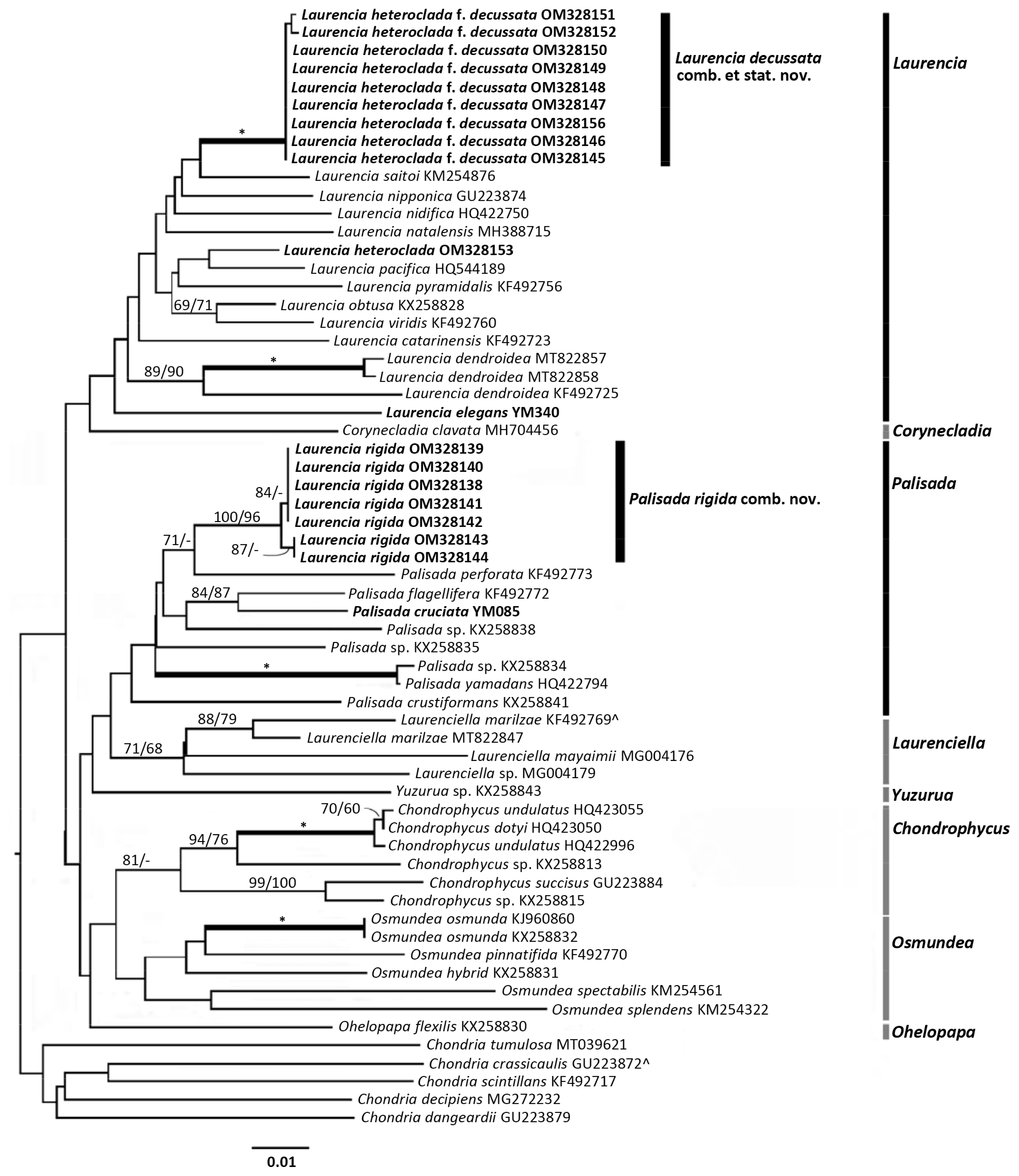
The neighbour-joining (NJ) tree based on COI-5P sequences data. Bootstrap values for NJ and maximum parsimony methods (1,000 replicates) greater than 70/70 are shown. Bold and * indicates full support. Sequences resulting from this study are in bold text, and those of topotype material are indicated with ^.
The L. rigida clade contained seven sequences. The clade grouped within the genus Palisada. Intraspecific p.d. within L. rigida were 0.00–0.35%. The closest neighbouring taxon was P. perforata with a pairwise distance of 5.11%.
The L. heteroclada f. decussata clade contained nine sequences. Pairwise distances within the clade were between 0.00–0.18%. The closest neighbouring sequence was L. saitoi with an interspecific pairwise distance between them of 3.52%.
The topotype sequence of L. heteroclada (GenBank No. OM328153) was included. As with the rbcL analyses, this sequence did not group near the L. heteroclada f. decussata clade. The interspecific distances between the two were 4.58–4.80%.
Morphology
To assist in distinguishing between L. rigida and L. heteroclada f. decussata, 16 morphological features were compared and recorded between these two taxa, as well as three other morphologically similar taxa. These are presented in Supplementary Table S2. These 16 morphological characters have been successfully used to separate between genera as well as species (Yamada 1931, Saito 1967, Saito and Womersley 1974, Garbary and Harper 1998, Nam 2006, Cassano et al. 2012, Metti et al. 2015).
Laurencia rigida showed no secondary pit connections between cortical cells when observed in longitudinal section. It also displayed perpendicular tetrasporangia alignment in relation to the main axial row. Laurencia rigida showed no corps en cerise, and had a small, discoid holdfast that was sometimes stoloniferous. Ultimate branchlets were very short when compared to the length of the supporting branch and were widely spread out from the supporting branch.
Laurencia heteroclada f. decussata showed parallel tetrasporangia alignment and the presence of secondary pit connections. In L. heteroclada f. decussata corps en cerise were seen, one each in surface and trichoblast cells. The holdfast was a densely tangled mass of stolons with multiple upright axes arising. The ultimate branchlets were pressed close to the supporting branch, forming a columnar outline. The ultimate branchlets were of moderate length when compared to the main axes and reproductive branchlets were often compound and densely grouped.
The rbcL and COI-5P results and morphological observations strongly support the transfer of L. rigida to Palisada as P. rigida comb. nov., and the raising of L. heteroclada f. decussata to specific rank as L. decussata comb. et stat. nov.
Taxonomic results
Laurencia decussata (A. B. Cribb) Metti comb. et stat. nov. (Figs 3–5)
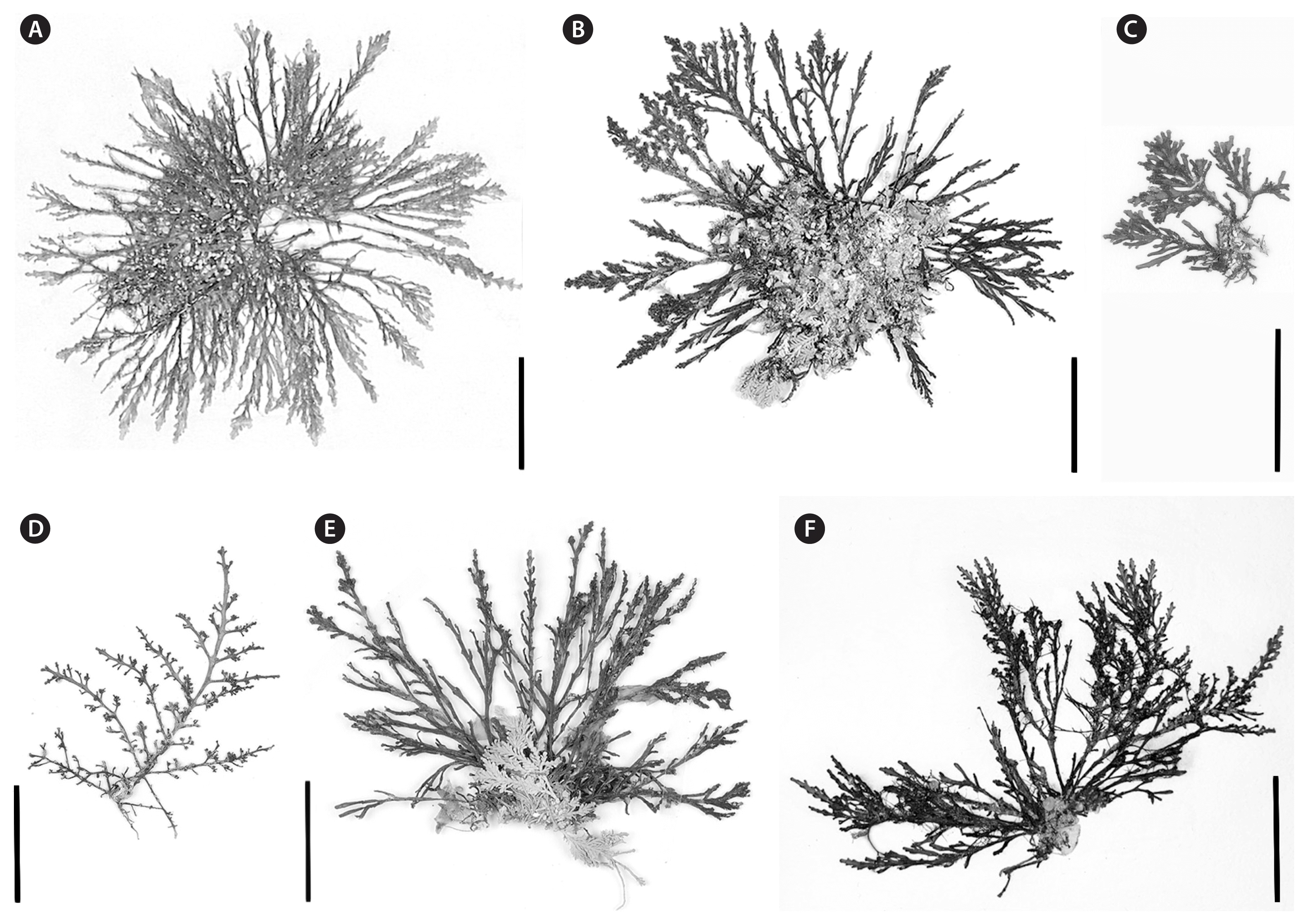
Plant habits of Laurencia decussata. (A) Isotype of Laurencia decussata (as Laurencia heteroclada f. decussata) labelled “BRI No.3.1 11.viii.1948”, collected by A. B. Cribb (Cribb 1958). (B) Habit of tetrasporic plant (NSW1115021), collected intertidally from northern NSW. (C) Habit of cystocarpic thallus (NSW1114926) collected intertidally from southern NSW. (D) A cystocarpic plant (NSW1114923) collected subtidally at 9 m, in southern NSW. (E) Habit of a sterile plant (NSW1115019) collected intertidally in northern NSW. (F) Male plant (NSW1114934) collected intertidally from North Stradbroke Island, Queensland, near the type locality. Scale bars represent: A–F, 2 cm.

Branching details of Laurencia decussata. (A) The lower portion of a whole plant detailing the stoloniferous holdfast (NSW857588). (B) The general branching pattern of a sterile plant showing both long and short ultimate branchlets pressed closely to the supporting axes (NSW857588). (C) Detail of cystocarpic branchlets and rounded cystocarps which lack protruding ostioles (NSW1114926). (D) The upper third of a tetrasporic branch, showing a crowded branching pattern (NSW1114939). Scale bars represent: A–D, 1 mm.

Microscopic features of Laurencia decussata. (A) Longitudinal section through a tetrasporic ultimate branchlet, showing parallel arrangement of tetrasporangia, undulating profile of branchlet and narrow apical pit (NSW1114939). (B) Both long and short trichoblasts emerge from apical pit (NSW857588). (C) Longitudinal section showing the undulating profile of a branchlet near the apical pit (NSW1115021). (D) Surface image showing one to two corps en cerise per epidermal cell (white arrow), and longitudinal secondary pit connections between surface cells (black arrows) (NSW857588). (E) Transverse section showing lenticular thickening (black arrow) and thickened cell walls (NSW1115021). Scale bars represent: A, 100 μm; B, C & E, 25 μm; D, 15 μm.
Basionym
Laurencia heteroclada Harvey f. decussata A. B. Cribb 1958, pp. 176–177, pl. 11, figs 1–3; pl. 12, figs 1–4.
Type locality
Miami, Queensland, Australia (Cribb 1958, p. 176).
Type specimens (Fig. 3A)
BRI No. 3.1, AQ712542, Australia, Queensland, Miami, Aug 11, 1948, 6 specimens (Cribb 1958).
Previous misapplied names for Australia
In A. J. K. Millar and G. T. Kraft 1993, p. 54 (as Laurencia filiformis (C. Agardh) Montagne 1845, p. 125). In A. J. K. Millar and G. T. Kraft 1993, p. 55, Millar 1990, p. 466, fig. 76a, Zuccarello and West 2006, p. 27, fig. 2 (as L. rigida J. Agardh 1876, p. 651). In J. A. Lewis 1984 (as L. filiformis f. heteroclada (Harvey) Saito and Womersley 1974, p. 834).
Diagnosis
With typical generic characters. Dark red to light orange-red to dark purple. Plant sizes range up to 7 cm tall, with heights commonly around 4.5 cm. In general, subtidal plants are larger. The holdfast is composed of a densely tangled mass of stolons from which multiple uprights arise, most being percurrent and frequently denuded or sparsely branched in the lower half. Plants forming clumps of many erect axes, on rock in lower intertidal to upper subtidal zones, commonly along wave-exposed shores. Branches are terete, fastigiate and often columnar, with ultimate branchlets pressed close to supporting axes, particularly in sterile or tetrasporic plants. Branching decussate, opposite or subopposite, rarely alternate. One corps en cerise per surface and trichoblast cells were seen. Cortical cells not protruding or only slightly at apices when viewed in longitudinal section. In cross section, four pericentral cells per vegetative axial segment are present, and few lenticular thickenings are seen in medullary cells. Tetrasporic branching extremely full at apex of supporting branch, with ultimate branches often compound and subcorymbose and pressed close to the supporting branch. Cystocarpic plants showing ovoid almost circular cystocarps, one per fertile branchlet. Branches of female plants often decussate, with more open angles between ultimate branchlets and the supporting axes. Branchlets on the female plant that are without cystocarps are often compound and sparsely spaced along main axes. Typically, main axes are around 1.5 mm in diameter. Male plants were up to 5.3 cm tall. Fertile branchlets ranged from 0.3 to 0.5 mm in length. Spermatangia developed from fertile trichoblasts within a cup-shaped apical pit and showed an apical nucleus within spermatia. The single terminal vesicle was sterile and obovate.
Habit
Most plants were dark red, but colours ranged from light orange-red to dark purple. Plant sizes ranged from 2–7 cm tall. In general, subtidal plants were larger. Plants when alive were sturdy and when pressed did not adhere well to paper. Fouling by other organisms was common, most samples being covered by some combination of other algae, diatoms, bryozoans, bivalves and isopods. The holdfast was composed of a densely tangled mass of stolons from which multiple uprights arose, most being percurrent. Branching orders ranged from three to five, but commonly plants displayed four branching orders. Branches were terete and often columnar. Plants found in NSW matched well with Cribb’s (1958) original description of the species from Queensland, although plants collected further south were generally slightly smaller.
Vegetative structures
In longitudinal sections, epidermal cells were oblong, and in transverse sections epidermal cells were isodiametric to inversely triangular. Secondary pit connections were seen between epidermal cells in longitudinal section. Epidermal cells were sometimes very slightly projecting at apices, but more often the branchlet showed only an undulating profile. One corps en cerise was seen in surface cells and in trichoblast cells, none were seen in medullary cells. Apical pits were shallow in relation to the size of the ultimate branchlets. Moderate amounts of trichoblasts were present within the pit. In transverse sections, few lenticular thickenings were present. Vegetative axial cells cut off four pericentral cells each.
Tetrasporic
Tetrasporic plants were the largest compared to other phases, with heights observed up to 7.0 cm. Tetrasporangial branchlets were often compound, numerous and densely grouped along the upper portions of the supporting branch. Angles between branches and branchlets were extremely tight, with ultimate ramuli pressed against the supporting branches. The sizes of tetrasporangial ultimate ramuli ranged from 0.5–3.0 mm long and 300–600 μm in diameter. Tetrasporangia showed parallel development in relation to the central axis and had a pericentral cell origin. They reached up to 144 μm in diameter and were connected to the supporting cell abaxially. Two fertile pericentral cells and two sterile ones were seen on fertile axes. Tetrasporangia scarring was observed on some branchlets.
Male
Male plants were up to 5.3 cm tall. Generally, habits resembled tetrasporic plants in branching patterns, with dense branching near the apical regions of axes. Up to four orders of branching were seen. Spermatangial ultimate branchlets were often compoundly branched, and showed a distinct widening at apices, resulting in a flattened pyriform shape. Fertile branchlets ranged from 0.3 to 0.5 mm in length. Spermatangia developed from fertile trichoblasts within a cup-shaped apical pit and showed an apical nucleus within spermatia. The single terminal vesicle was sterile and obovate.
Female
Cystocarpic plants were up to 5.7 cm tall and were collected both intertidally and subtidally down to 9 m. Up to four branching orders for subtidal specimens were observed. The ultimate branchlets were evenly distributed along secondary and tertiary branches at 45-degree angles or more. Fertile branchlets were sometimes compound but most often they were single and bearing one cystocarp each. Cystocarps were located at the upper third of branches and were widely ovate, almost circular, with ostioles that did not protrude. Cystocarps were seen up to 0.75 mm long and 0.60 mm in diameter. Carpospores are generally lanceolate, with a maximum observed length of 207 μm and diameter of 39 μm.
Specimens examined
The type specimens BRI AQ712542; NSW1114915, collector number YM047 and NSW1114916, collector number YM049, collected Jul 25, 2004, Australia, NSW, Emerald Beach (30°10′17.9″ S, 153°11′27.9″ E); NSW857588, collector number YM069 sterile, and NSW1114918, collector number YM070, collected Jul 28, 2004, Australia, NSW, Arrawarra Headland (30°03′32.3″ S, 153°12′19.4″ E); NSW1115019, collector number YM072, sterile, collected Jul 28, 2004, Australia, NSW, Arrawarra Headland (30°03′32.3″ S, 153°12′19.4″ E); NSW1114921, collector number YM139, collected Feb 8, 2005, Australia, NSW, Newcastle, Newcastle harbour (32°55′24.4″ S, 151°47′20.5″ E); NSW1114922, collector number YM153, collected Feb 9, 2005, Australia, NSW, Sydney, Bare Island (33°59′27.9″ S, 151°13′55.1″ E); NSW1114923, collector number YM171 and NSW1114924, collector number YM178, collected Feb 15, 2005, Australia, Jervis Bay, Plantation Point (35°04′16.1″ S, 150°41′48.8″ E); NSW1114926, collector number YM212, cystocarpic, collected Feb 16, 2005, Australia, Jervis Bay, Plantation Point (35°04′16.1″ S, 150°41′48.8″ E); NSW1115021, collector number YM392, tetrasporic, collected Aug 29, 2006, Australia, NSW, Arrawarra Headland (30°03′32.3″ S, 153°12′19.4″ E); NSW1114939, collector number YM393, tetrasporic, collected Aug 29, 2006, Australia, NSW, Arrawarra Headland (30°03′32.3″ S, 153° 12′19.4″ E); NSW1114934, collector number YM779, tetrasporic, collected Oct 7, 2018, Australia, Queensland, North Stradbroke Island, Adder Rock Beach (27°25′15.9″ S, 153°30′54.8″ E).
Australian distributions
From North Stradbroke Island, Queensland, down the east coast to Tathra, NSW.
Habitat and seasonality
Laurencia decussata was collected throughout NSW and in southern Queensland. It was collected from the lower intertidal down to 9 m subtidal, always on large rocky substrates such as rock shelves or very large boulders on wave-exposed shores or within turbulent waters. No plants found were epiphytic. Tetrasporic material was collected in the winter, spring and summer. Carposporic plants were collected in summer, and sterile material was collected in winter. Spermatangial material was found in spring.
Palisada rigida (J. Agardh) Metti comb. nov. (Figs 6 & 7A–D)
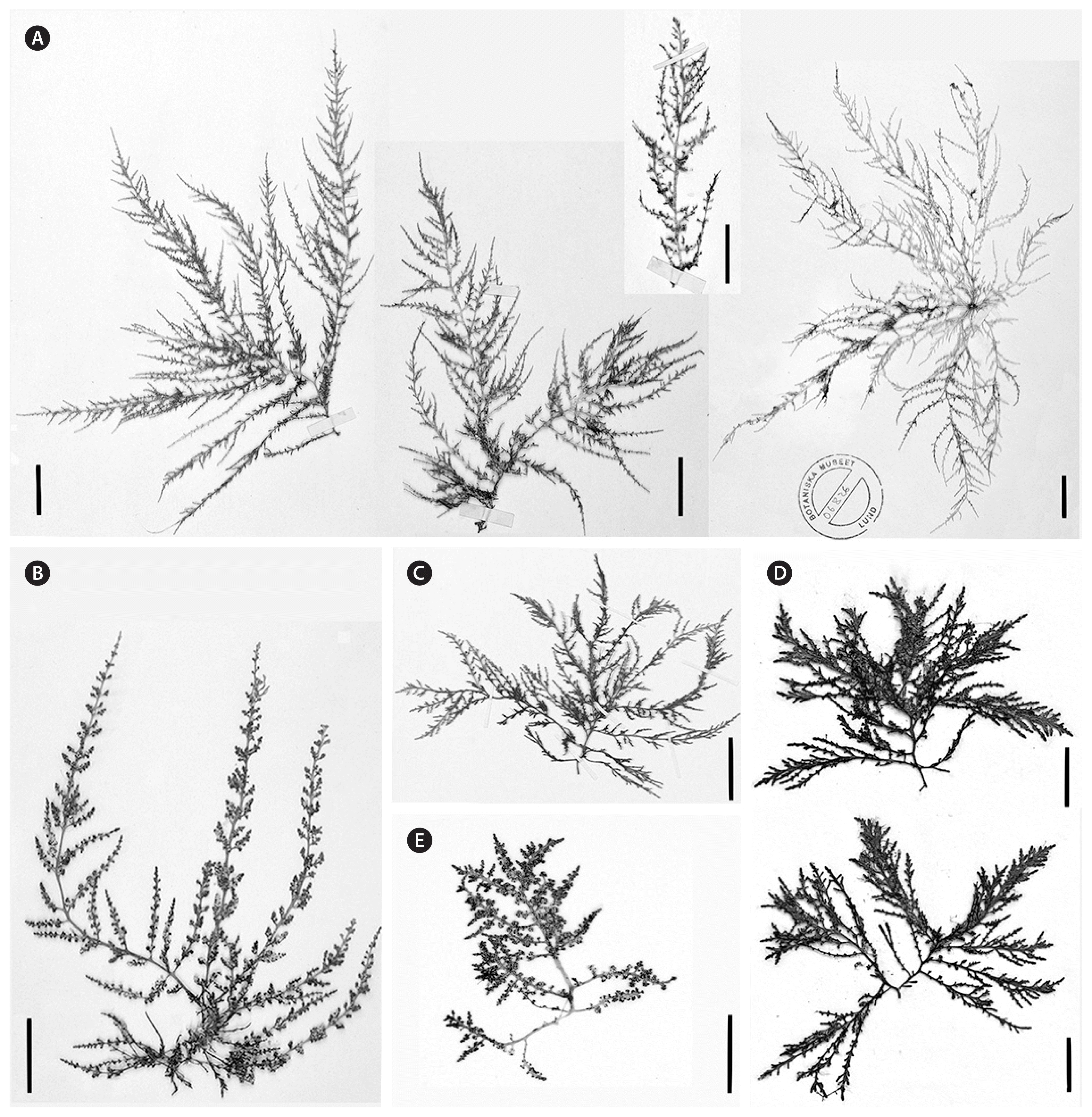
Palisada rigida plant habits. (A) Type specimen (as Laurencia rigida), from J. Agardh (1876), including all four specimens from the original sheet labelled “LUND 36694, Laurencia rigida J. Ag. e Nova Hollandia boreali” collected by Kilner. (B) Palisada rigida (as Laurencia papillosa var. 2, later Laurencia rigida) collected by Harvey and labelled as “LUND 36695. Laurencia papillosa var. 2, 240D Harvey Austral. Alge.” This is also incorrectly labelled as “TYPUS.” (C) Isotype specimen (as Laurencia rigida) also collected by Kilner and labelled “MEL1007183, Laurencia rigida J. Ag., Nov. Holl. boreal.” (D) Newly collected tetrasporic specimens (NSW1114937) from Redcliffe, Queensland, which is near the assumed type locality. (E) Habit of a well developed tetrasporic plant (NSW1114927) collected intertidally from southern NSW. Scale bars represent: A–E, 2 cm.
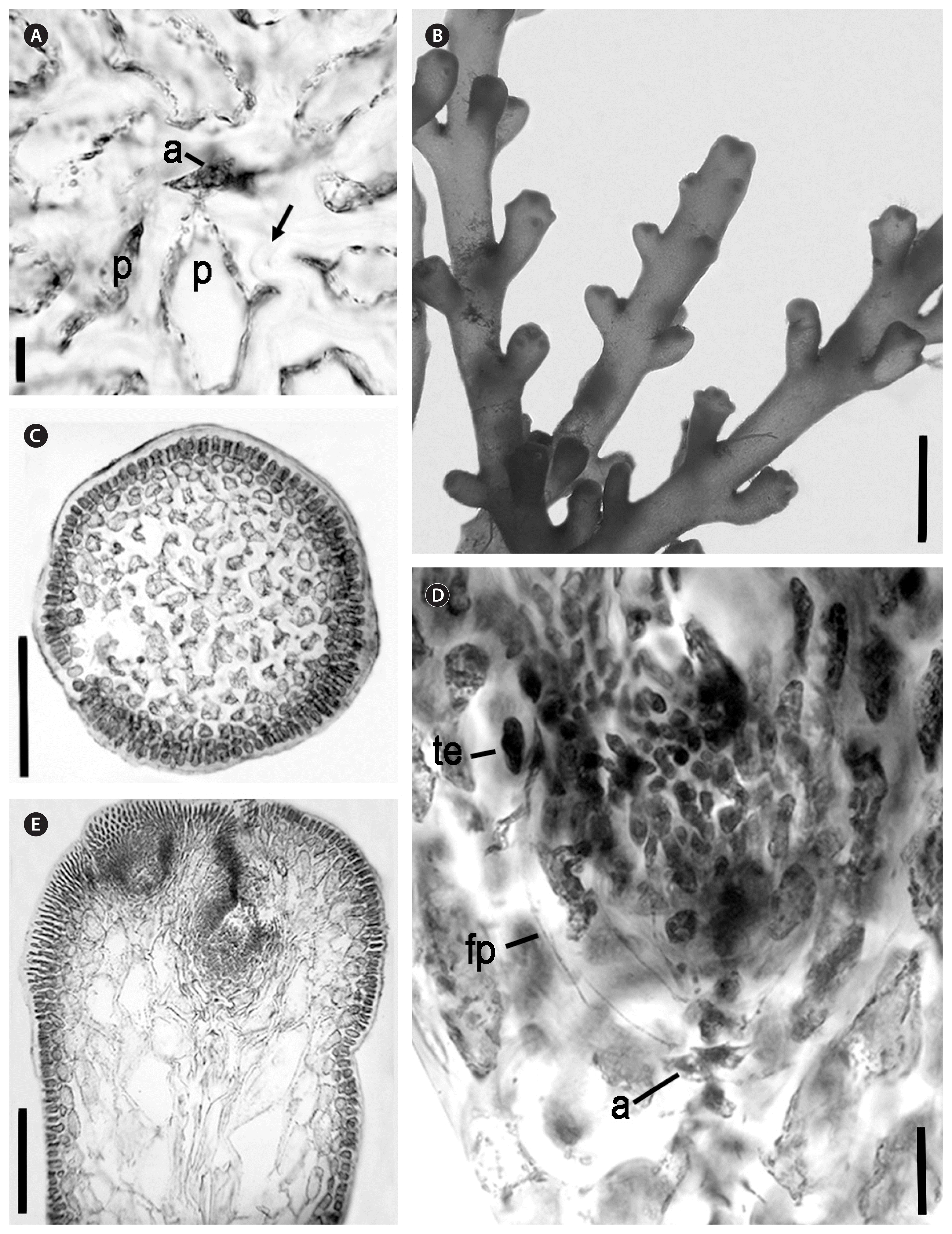
Microscopic features of Palisada rigida. (A) Transverse section showing central axial cell (a) with two pericentral cells (p) and thickened cell walls (arrow) (NSW1115020). (B) The upper portion of branches (NSW1114937) showing the ultimate branchlets inserted into the supporting branch at close to a 45-degree angle, and a tapering profile towards the apex. (C) Transverse section showing palisade-like formation of outer cortical layer (NSW1115020). (D) Longitudinal section through a tetrasporangial branchlet (NSW1114931) showing the tetrasporangium (te) developing from a fertile pericentral cell (fp), and the axial cell (a). (E) Longitudinal section showing the profile of an ultimate branchlet (NSW1115020). Scale bars represent: A, 100 μm; B, 6 mm; C & E, 200 μm; D, 25 μm.
Basionym
Laurencia rigida J. Agardh 1876, p. 651.
Type locality
“in oceano indico calidiore” from the warmer parts of Australia (J. Agardh 1876, p. 651).
Type specimens (Fig. 6A)
Agardh LUND #36694, “e Nova Hollandiea boreali; Kilner”. Probable Isotype specimens (Fig. 6C): Mueller, MEL 1007183 “nov. Holi. boreal; Kilner”; BM 974110 “Nov. Holi. boreal.”; BM 974113 “nov. Holi, boreal.”
Diagnosis
With typical generic characters. Plant colours range from dark red to brown to light green. Plant heights range from 2.2–13.7 cm. The thallus is terete throughout with one or more percurrent axes. The holdfast is small and discoid, sometimes slightly stoloniferous. Branches are sturdy and cartilaginous, displaying sparse, ternate branching both alternate and often opposite. Up to four orders of branching are seen but commonly only three are present. Angles between branches are between 45 to 90-degrees. No secondary pit connections between epidermal cells are seen in longitudinal sections. In cross section, epidermal cells are slightly palisade. Epidermal cells are not projecting. Two pericentral cells per vegetative axial segment were observed. There were no lenticular thickenings found, although thickened cell walls were seen. No corps en cerise observed. Tetrasporic plants show fertile ultimate branchlets that are compound and forming botryoidal clusters that were regularly distributed along supporting branch. Angles are often close to 90-degrees between tetrasporic cluster and supporting branches. Cystocarpic plants show ultimate branchlets slightly longer than other phases. Cystocarps are widely ovoid with protruding ostioles. They develop embedded in the supporting branches, in between ultimate ramuli and close to branch apices.
Habit
Plants showed a terete habit. Most plants showed to be dark red or brown, with some showing lighter colour or even green. Pressed plants adhered well to paper. Fouling was generally low or not existant. The holdfast was discoid, often a single upright arose, but it was not uncommon to see multiple uprights. Generally three or sometimes four branching orders were seen. Main axes ranged from 0.7–1.2 mm in width. The ultimate branchlets are very short in comparison to their supporting branch and inserted into the supporting branch at a wide angle. The ultimate branchlets shorten along the length of the supporting branch towards the apex, creating a long, gradual tapering outline. This pattern is repeated at each branching order.
Vegetative structures
Secondary pit connections were absent between epidermal cells in longitudinal section. Corps en cerise were also absent from any cells, as were lenticular thickenings. However, some cell wall thickenings were observed. Epidermal cells were not projecting at apices, but often showed a palisade-like structure in cross section. A moderate number of trichoblasts were present within the pit. Vegetative axial cells cut off two pericentral cells each.
Tetrasporic
Tetrasporic plants reached upto 13.7 cm tall, with the average observed height at 7.8 cm. Fertile ultimate branchlets narrowed at the apex of the branchlet, and were at an average length of 0.4 mm long. They were compound and formed botryoidal clusters. These clusters were often distributed regularly along the supporting branch. Clusters ranged in sizes from 0.7 mm wide × 0.6 mm long to 1.7 mm wide × 1.2 mm long. Angles between tetrasporic clusters and their supporting branch were often almost perpendicular. Tetrasporangial arrangement when observed in longitudinal section was perpendicular to the main axial row with a pericentral cell origin. One sterile pericentral cell and one fertile pericentral cell were present on the tetrasporangial axis. Tetrasporangia reached up to 100 μm in diameter.
Male
Male plants were not observed.
Female
Female plants averaged 7.5 cm in height with the largest height observed at 13 cm and the shortest was 4.9 cm. In general, plants of all phases observed were generally of the same size. However, female plants were more often seen with multiple uprights arising from the holdfast, in comparison to other phases. The ultimate branchlets in cystocarpic plants were slightly longer than other phases as well. Cystocarps were widely ovoid with definite, protruding ostioles. Cystocarps measured upto 1.4 mm in length, and the protruding ostiole measured upto 400 μm long. Cystocarps developed in between ultimate branchlets, close to the supporting branch apices.
Specimens examined
Harvey LUND #36695, Harvey Austral. Algae; MEL 1007183, collector is Kilner, F., identified by J. Agardh, from “Nov. Holl. boreal.”; Agardh LUND #36695, “Nova Hollandia boreali”; Lucas NSW, Botany Bay, Feb 1905, #1–7; Lucas NSW, Lake Macquarie, Jan 1918, #1 and #2; NSW290986, Feb 7, 1992, Australia, NSW, 700 m West North West of Mowarry Point, South of Twofold Bay, (37°08′20″ S, 149°59′45″ E); NSW1114937, collector number Q014, May 30, 2013, Australia, Queensland, Redcliffe (27°13′59.7″ S, 153°07′03.7″ E); NSW1114925, collector number YM208 and NSW1114927, collector number YM219, Feb 16, 2005, Australia, Jervis Bay, Plantation Point (35°04′16.1″ S, 150°41′48.8″ E); NSW1114928, collector number YM230 and NSW1114929, collector number YM234, Mar 15, 2005, Australia, Norfolk Island, Kingston Lagoon, Slaughter Bay (29°03′32.5″ S, 167°57′27.7″ E); NSW1115020, collector number YM382, Nov 17, 2005, Australia, NSW, Botany Bay, Kurnell (34°00′27.2″ S, 151°12′24.9″ E); NSW1114931, collector number YM385, Nov 17, 2005, Australia, NSW, Botany Bay, Kurnell (34°00′27.2″ S, 151°12′24.9″ E).
Australian distributions
From Cape Upstart, just south of Townsville in Queensland, down the east coast throughout NSW and to Port Fairy, just west of Warnambool in Victoria, including Norfolk Island and Lord Howe Island.
Habitat and seasonality
Palisada rigida was found intertidally in tide pools on rock platforms, often with sand or gravel present. It was also found subtidally down to 21 m, on rock or coral reefs surrounded by sand. Collections were made in every season. Slight fouling was seen, often by Ceramium sp., Gelidium sp., worms, diatoms, brown and green filamentous algae.
DISCUSSION
Many herbarium specimens labelled as “Laurencia rigida” from various Australian herbaria on careful inspection seemed to more closely resemble the type of Laurencia decussata (as L. heteroclada f. decussata). During morphological examinations of both Palisada rigida (as L. rigida) and L. decussata (as L. heteroclada f. decussata), three generic characters and four specific characters presented themselves as most useful in separating between them. The three generic characters were; number of pericentral cells per vegetative axial segment, the presence or absence of secondary pit connections between cortical cells as seen in longitudinal section, and parallel or perpendicular tetrasporangia alignment when compared to the main axial row. These three characters are commonly used to separate taxa between the Laurencia, Laurenciella, and Corynecladia genera from the Palisada and Chondrophycus genera (Nam 2006, Cassano et al. 2012, Metti et al. 2015, Rousseau et al. 2017). Palisada rigida showed; two pericentral cells per vegetative axial segment, no secondary pit connections, and a perpendicular tetrasporangia alignment when compared to the main axial row. Laurencia decussata on the other hand showed; four pericentral cells per vegetative axial segment, a parallel tetrasporangia alignment, and the presence of secondary pit connections. The two taxa showed features typical of their genus (Saito 1967, Saito and Womersley 1974, Nam 2006, 2007, Metti et al. 2015, Rousseau et al. 2017). The morphological characters of secondary pit connections and tetrasporangia alignment are commonly used and relatively simple to observe, and therefore are useful as a first step in separating the two taxa.
Four other morphological characters were also useful in distinguishing between P. rigida and L. decussata. These included; the presence or absence of corps en cerise, the presence or absence of lenticular thickenings within medullary cells, holdfast morphology, and overall plant shape. When looked at in combination they can be useful in distinguishing between these two species, particularly with pressed specimens. Corps en cerise are present only in Laurencia and Laurenciella. They are present in all species of these two genera, but visible only in living or freshly collected material. Lenticular thickenings are present only in Laurencia and Osmundea, but it is a highly variable feature among species. Palisada rigida showed no corps en cerise, and no lenticular thickenings. It had a small, discoid holdfast that was sometimes stoloniferous. Ultimate branchlets were very short, were inserted into the supporting branch at wide angles and distributed somewhat evenly along the length of the supporting branch. Branching profiles often showed a long tapering towards the apex. However, in L. decussata lenticular thickenings were seen, as were corps en cerise, one each in surface and trichoblast cells. The holdfast was a densely tangled mass of stolons with multiple upright axes arising. Reproductive branchlets were often compound and densely grouped, particularly at the apical end of the supporting branch. Laurencia decussata had robust and often decussate branching with ultimate ramuli pressed close to the supporting branch, whereas the type of P. rigida had much longer branches overall, except for the ultimate ramuli which were shorter but less densely branched. The main branches in L. decussata were mostly denuded near the base and more profusely branched at the apical end, whereas in P. rigida the basal branches were generally longer than those near the apex.
Palisada rigida when tetrasporic can also superficially resemble both Laurencia botryoides and L. arbuscula. However, besides showing typical Laurencia genus features such as the presence of secondary pit connections, both L. arbuscula and L. botryoides showed a densely stoloniferous holdfast. In contrast, P. rigida showed Palisada genus features such as the absence of secondary pit connections and perpendicular tetrasporangia alignments, as well as a small, discoid holdfast, which rarely developed supporting stolons. Laurencia botryoides showed very short ramuli grouped in tight clusters whether tetrasporic or not. It often had regular, distichous, alternate branching. This was most evident along the percurrent axes. It also had thick and sturdy primary axes. However, in P. rigida the ultimate ramuli were short, widely and evenly spaced, and usually single or in small, loose clusters when tetrasporic. Palisada rigida developed alternate to opposite branching, lacking the clearly visible pattern that was often seen in L. botryoides. On the other hand, Laurencia arbuscula had ultimate branches that were generally longer than those in P. rigida. They were primarily fastigiate and grouped at the apex of the supporting branch. Laurencia arbuscula showed opposite to alternate branching patterns as well, but in P. rigida the secondary branches were generally as long or longer than the percurrent axes, which was not the case with L. arbuscula.
In J. Agardh’s LUND collection, two specimens of P. rigida (as L. rigida) are labelled as “TYPUS”; LUND #36695, which is a Harvey specimen, and LUND #36694, which is the Agardh specimen. The correct type of L. rigida is the specimen from northern Australia (LUND #36694) as was stated in J. Agardh (1876) (Yamada 1931, Millar 1990). It is not the Harvey collected specimen from Port Fairy in Victoria (LUND #36695). Although at quick glance these two specimens looked to be separate taxa, the Harveyan specimen is a fertile form of P. rigida. Agardh (1876) himself included the Harveyan specimen (L. papillosa var.2 Harvey Alg. Austr. exs. #240) in his protologue of L. rigida. Morphological comparisons of the habits of both supported these two specimens belonging to the same species. In particular, they shared similar habit shape, including a distinct, long narrowing of the branch apices. They showed similar branching structures, both having longer secondary branches closer to the base of the plant, and very short ultimate branchlets spread out along the supporting axes. The obvious morphological differences between these two primarily stem from the Harveyan specimen being tetrasporic. As is commonly seen within the Laurencia complex, tetrasporic plants show slightly thicker and many more ultimate branchlets than other phases (Metti et al. 2015).
There is a specimen in MEL (MEL1007183) from the Algae Muellerianae, collected in “Nov. Holi. boreal; Kilner”, that is very likely an isotype of P. rigida (as L. rigida). It morphologically matched well with the Agardh type 36694, although it was smaller. This specimen was annotated by Yuzuru Saito as L. rigida, dated Apr 12, 1972, that “no pit-connections between each epidermal cell; right angle type arrangement in tetrasporangia, L. rigida should be a member of subgen. Chondrophycus.” This was previous to the establishment of Palisada. The molecular analyses from this study supports Saito’s observation that L. rigida does not belong in the Laurencia genus. In fact, molecular results showed specimens that morphologically match the type specimens to be in the genus Palisada. Within the molecular analyses there was a sequence (GenBank No. OM328131) included from a newly collected specimen from Queensland (NSW1114937) that morphologically matches well with MEL1007183, the isotype of P. rigida (as L. rigida). In particular, they both show long, tapered branch profiles, short ultimate ramuli and a small discoid holdfast. Both the Queensland specimen and the isotype seem to be slightly smaller but still well matched morphologically with the Agardh type LUND36694. In the molecular results for both rbcL and COI-5P this newly collected Queensland specimen sits within the genus Palisada, along with nine other newly collected sequences.
A newly collected specimen of L. decussata was also collected from Queensland. This specimen morphologically matched very well with the type specimens of L. heteroclada f. decussata, including a well developed, stoloniferous holdfast with multiple, columnar uprights. All the newly collected specimens that morphologically matched the type formed a monophyletic clade in all molecular analyses within the Laurencia genus. One sequence included in the L. decussata clade was downloaded from GenBank and is labelled “Laurencia rigida” (GenBank No. AY920852). Unfortunately, the voucher has not been located, however it is a specimen from NSW where L. decussata was commonly identified as “L. rigida” and is most likely based on a misidentified specimen.
Laurencia decussata was originally described as a variety of L. heteroclada. Included in these analyses was a sequence of L. heteroclada (GenBank No. OM328153) that was collected from the type locality of Rottnest Island, WA. Morphologically it matched well with the L. heteroclada type (TCD0015207, Herb. Harvey Trav. Set No. 210). This topotype sequence of L. heteroclada did not group closely to the L. decussata clade in either rbcL or COI-5P analyses. Morphologically the two taxa can be separated by L. heteroclada having ultimate branchlets that are long and inserted at a wide angle into the supporting branch, whereas in L. decussata the ultimate branchlets are generally shorter, and usually pressed close to their supporting branch. The lower order branches are often longer in L. heteroclada than in L. decussata when compared to the overall plant size. Laurencia decussata also generally has thicker primary axes, and a more developed, stoloniferous holdfast. One reason for Cribb (1958) separating the two taxa was their slightly disjunct distributions within Queensland. He noted that L. heteroclada was found north of Redcliffe, and L. decussata (as L. heteroclada f. decussata) occurred south of Miami (Cribb 1958). This still seems to be the case. Currently L. decussata (as L. heteroclada f. decussata) is still only recorded from NSW and southern Queensland (Cribb 1958), and L. heteroclada is found in WA, South Australia, Victoria, northern Tasmania, and then continues from Redcliffe in Queensland northwards (Cribb 1958, Saito and Womersley 1974).
In the molecular analyses, rbcL showed good resolution for species and genera in the Laurencia complex. COI-5P on the other hand did not resolve the generic clades within this group. However, even with lack of support for the generic clades in COI-5P, the results for the P. rigida and L. decussata clades were well supported with an obvious separation seen between P. rigida and L. decussata, as well as between L. decussata and L. heteroclada.
ACKNOWLEDGEMENTS
This work was supported by the Australian Biological Resources Study (ABRS) under Grant RFL212-08, which funded a large portion of the field and molecular work. This study was also supported by the National Herbarium of NSW, Sydney, Australia. Thank you to J. Eu, P. Ogilvie, A. J. K. Millar, S. Skinner and D. Ghosn for assisting in collecting specimens. Thank you also to the reviewers of this manuscript for their detailed comments.
Notes
The authors declare that they have no potential conflicts of interest.
SUPPLEMENTARY MATERIALS
Supplementary Table S1. A list of species used for molecular work in this study, including newly generated rbcL and COI-5P sequences, and downloaded GenBank sequences (https://www.e-algae.org).
Supplementary Table S2. Comparison of morphological features between Laurencia rigida, L. heteroclada f. decussata, and three other morphologically similar taxa (https://www.e-algae.org).