Spatial-temporal distributions of the newly described mixotrophic dinoflagellate Gymnodinium smaydae in Korean coastal waters
Article information
Abstract
Gymnodinium smaydae is a newly described mixotrophic dinoflagellate that feeds on only Heterocapsa spp. and Scrippsiella acuminata among 19 tested algal prey. It is one of the fastest growing dinoflagellates when feeding, but does not grow well without prey. To investigate its spatial-temporal distributions in Korean waters, we quantified its abundance in water samples that were seasonally collected from 28 stations along the Korean Peninsula from April 2015 to October 2018, using quantitative real-time polymerase chain reactions. This dinoflagellate had a wide distribution, as reflected by the detection of G. smaydae cells at 23 of the sampling stations. However, this distribution had a strong seasonality; it was detected at 21 stations in the summer and only one station in winter. The abundance of G. smaydae was significantly and positively correlated with chlorophyll a concentration as well as with water temperature. However, there were no significant correlations between the abundance of G. smaydae and salinity, concentrations of nutrients, or dissolved oxygen concentration. During the study period, G. smaydae was present when water temperatures were 7.6–28.0°C, salinities were 9.6–34.1, concentrations of NO3 were not detectable-106.0 μM, and concentrations of PO4 were not detectable-3.4 μM. The highest abundance of G. smaydae was 18.5 cells mL−1 in the coastal waters of Jinhae in July 2017 when the chlorophyll a concentration was 127 mg m−3 and water temperature was 23.8°C. Therefore, the spatial-temporal distribution of G. smaydae in Korean coastal waters may be affected by chlorophyll a concentration and water temperature.
INTRODUCTION
Dinoflagellates are ubiquitous and major components of marine ecosystems (Smayda 1997, Sherr and Sherr 2007, Taylor et al. 2008, Jeong et al. 2010, 2015, Lim et al. 2017). They display all three trophic modes: autotrophic, heterotrophic, and mixotrophic (Menden-Deuer et al. 2005, Burkholder et al. 2008, Hansen 2011, Jeong et al. 2016, 2018, Stoecker et al. 2017, Lim et al. 2019a). Over the last two decades, several mixotrophic dinoflagellates have been newly described (Yoo et al. 2010, Kang et al. 2011, Jeong et al. 2012, Lim et al. 2015, Jang et al. 2017, Yokouchi et al. 2018) and many dinoflagellates that were thought to be exclusively autotrophic have been revealed to be mixotrophic (Stoecker 1999, Johnson 2015, Jeong et al. 2016, Millette et al. 2017, Lim et al. 2018, Yoo et al. 2018). Mixotrophic dinoflagellates play diverse roles in marine food webs as primary producers, prey, predators, symbionts, and parasites (Hansen 1992, Coats 1999, Jeong 1999, Skovgaard et al. 2012, Caron 2016, Gómez and Gast 2018, Flynn et al. 2019, Kang et al. 2019a). Many mixotrophic dinoflagellates are known to cause red tides or harmful algal blooms, which cause severe mortalities of fishes and shellfishes (Burkholder et al. 1995, Glasgow et al. 1995, Yoo et al. 2009, Jeong et al. 2017, Flynn et al. 2018, Shumway et al. 2018). Therefore, the distribution of mixotrophic dinoflagellates is important in understanding the structure and function of marine planktonic communities and to minimize losses due to red tides or harmful algal blooms (Burkholder et al. 1995, Taylor et al. 2008, Kudela and Gobler 2012, Flynn et al. 2018, 2019, Lee et al. 2018, 2020).
The distribution of mixotrophic dinoflagellates is known to be affected by biotic factors such as prey availability, predators, and competitors, and abiotic factors such as temperature, light intensity, and nutrient concentrations (Bockstahler and Coats 1993, Jacobson and Anderson 1996, Skovgaard 1996, Stoecker et al. 1997, Adolf et al. 2006, Burkholder et al. 2008, Hansen 2011, Kang et al. 2011, Lee et al. 2014b, Lim et al. 2015, Millette et al. 2017, Yoo et al. 2018). The distribution of a mixotrophic dinoflagellate is a result of a combination of these critical biotic and abiotic factors (Jeong et al. 2015). Therefore, analyses of the abundances of the target mixotrophic dinoflagellate and its prey as well as of abiotic factors in waters collected at many locations at different times are needed (Burkholder et al. 1995, Jeong et al. 2017, Lee et al. 2017, 2019, Kang et al. 2019b, Jang and Jeong 2020).
Quantification of the abundance of mixotrophic dinoflagellates using microscopy is not easy if the sizes of the mixotrophic dinoflagellates are <20 μm and there are sister species with similar morphologies (Lim et al. 2015, Lee et al. 2017, 2018, 2019, LaJeunesse et al. 2018, Kim et al. 2019, Jang and Jeong 2020). To solve this problem, the quantitative real-time polymerase chain reaction (qPCR) method has been used to quantify abundances of small mixotrophic dinoflagellates in field samples (Smith et al. 2016, Lee et al. 2017, 2018, 2019, Kang et al. 2019b). However, when using this method, a species-specific primer and probe should be developed for each species and a standard curve of fluorescence against cell abundance (DNA amount) using a pure culture of the target species must be established (Lee et al. 2017).
The dinoflagellate Gymnodinium smaydae, belonging to the family Gymnodiniaceae, was newly described in 2014 (Kang et al. 2014). This dinoflagellate was revealed to be a mixotrophic species (Lee et al. 2014a, Jeong et al. 2018, You et al. 2020). In a previous study, it was shown to only be able to feed on the dinoflagellates, Heterocapsa spp. (H. rotundata, H. steinii, and Heterocapsa sp. CCMP3244) and Scrippsiella acuminata, among 19 tested algal species (Lee et al. 2014a, Jeong et al. 2018, You et al. 2020). Its maximum growth rate is the highest among mixotrophic dinoflagellates (Berge et al. 2008, Yoo et al. 2010, Jeong et al. 2011, 2012, Kang et al. 2011, Lee et al. 2014a). However, it can grow well when fed on prey and does not grow without prey (Lee et al. 2014a, You et al. 2020), indicating that its growth rate may be primarily affected by prey availability. In addition, in a previous study, the growth rate of G. smaydae was also significantly affected by light intensity and water temperature (You et al. 2020). Some heterotrophic protists such as the common heterotrophic dinoflagellates Oxyrrhis marina and Gyrodinium dominans and the naked ciliate Pelagostrobilidium sp. are able to feed on G. smaydae, but are not preferred prey for these predators (Jeong et al. 2018). Therefore, based on the results of previous laboratory experiments, the population dynamics of G. smaydae can be suggested to be affected by prey availability, water temperature, and light intensity. The spatial and temporal distribution of G. smaydae could be the result of a combination of these environmental factors, and thus it is worthwhile to explore its spatial and temporal distribution.
In this study, to investigate the spatial-temporal distributions of G. smaydae, water samples were collected seasonally from 28 stations in the East, West, and South Seas of Korea and Jeju Island from April 2015 to October 2018. The size of G. smaydae (equivalent spherical diameter = 10.6 μm) is small, and thus the abundance of G. smaydae was quantified using qPCR. To use this method, a species-specific primer and probe for this species was developed and a specificity test conducted with this species as well. Furthermore, a standard curve of fluorescence against cell abundance (DNA amount) was established using pure culture of G. smaydae. Data on the physical and chemical properties of water, such as water temperature, salinity, nutrient concentrations, dissolved oxygen (DO) concentration, and chlorophyll-a (Chl-a) concentration from all sampled stations were obtained from our previous studies (Lee et al. 2018). The results of this study provide a basis for understanding the distribution of G. smaydae and critical environmental factors affecting this distribution.
MATERIALS AND METHODS
Collection of field samples
Field samples of G. smaydae were collected from the surface waters of 28 stations located in the East, West, and South Seas, as well as at Jeju Island, in Korea using a clean bucket in April, July, and October of 2015; January, March, July, October, and December of 2016; March, July, and October of 2017; and January, March, July, and October of 2018 (Fig. 1). The physical and chemical properties of surface waters at these stations were obtained from our previous studies (Lee et al. 2018, 2019, Kang et al. 2019b, Jang and Jeong 2020).
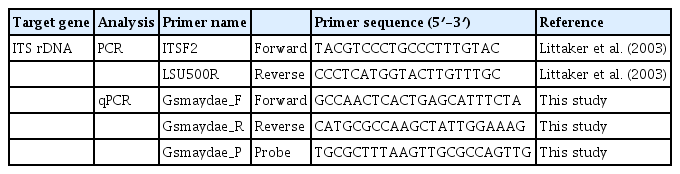
Map showing the sampling stations and distribution of Gymnodinium smaydae in Korean coastal waters during the study period. Red closed circles indicate stations at which G. smaydae cells were detected, whereas black open circles indicate stations at which G. smaydae cells were not detected. SC, Sokcho; JMJ, Jumunjin; DH, Donghae; UJ, Uljin; PH, Pohang; US, Ulsan; BS, Busan; DDP, Dadaepo; MS, Masan; JH, Jinhae; TY, Tongyeong; YS, Yeosu; KY, Kwangyang; GH, Goheung; JAH, Jangheung; AS, Ansan; DAJ, Dangjin; MGP, Mageompo; SCN, Seocheon; KS, Kunsan; BA, Buan; MP, Mokpo; AW, Aewol; GS, Gosan; SGP, Seogwipo; WM, Wimi; SS, Seongsan; GN, Gimnyeong.
Culture of Gymnodinium smaydae
A clonal culture of G. smaydae GSSH1005, originally isolated from Shiwha Bay, Korea, was used for qPCR (Kang et al. 2014). The culture was maintained in 250-mL PC bottles filled with freshly filtered and autoclaved seawater and H. rotundata as prey. The bottles were placed on a shelf at 20°C and illuminated with an irradiance of 20 μmol photons m−2 s−1 provided by a cool white fluorescent light under a 14 : 10-h light-dark cycle.
Species-specific primers and probe design, and specificity analysis
The internal transcribed spacer ribosomal DNA sequences of G. smaydae were aligned with sequences of other Gymnodinium species and related dinoflagellates, which were available from GenBank, using the program MEGA v. 4 (Tamura et al. 2007). Manual searches of the alignments were conducted to find unique sequences for developing a G. smaydae-specific qPCR assay. The sequences for the primer-probe set were selected from the regions that were conserved in G. smaydae but allowed for discrimination with other dinoflagellates. Primers and probe sequences of G. smaydae were analyzed using Primer 3 (Whitehead Institute, Cambridge, MA, USA; Howard Hughes Medical Institute, Chevy Chase, MD, USA) and Oligo Calc: Oligonucleotide Properties Calculator (Kibbe 2007) software for determining the optimal melting temperature and secondary structure. Subsequently, primers and probe were synthesized by Bioneer (Daejeon, Korea). The probe was dual-labeled with the fluorescent dyes FAM and BHQ1 (Bioneer) at the 5′ and 3′ ends, respectively (Table 1). To validate the specificity of specific primers and probe for G. smaydae, a specificity analysis was conducted using DNA extracts of other Gymnodinium species, related species, and prey species of G. smaydae (Table 2).
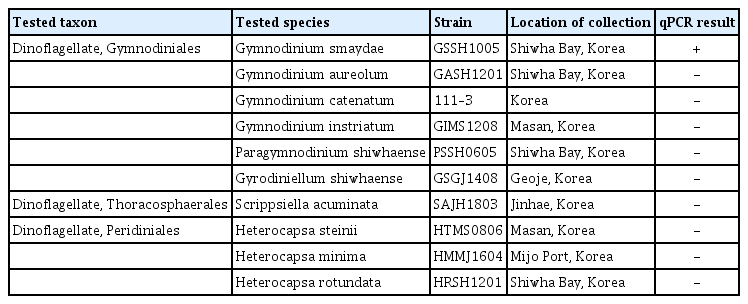
List of species used to determine primer specificity for the detection of Gymnodinium smaydae and quantitative real-time polymerase chain reaction (qPCR) results
qPCR to test the specificity of primers and probe set was performed using the following steps modified from Lee et al. (2018): 1 μL of DNA template, 0.2 μM of specific primers (forward and reverse), 0.15 μM of specific probe (final concentrations), and 5 μL of qPCRBIO Probe Separate-ROX (Genepole, Gwangmyeong, Korea) were combined, and deionized sterilized water (DDW) was added to each sample, resulting in a total final volume of 10 μL. qPCR assay was performed using Rotor-Gene Q (Qiagen, Hilden, Germany) under the following thermal cycling conditions: 3 min at 95°C, followed by 45 cycles of 10 s at 95°C, and 40 s at 58°C.
Generation of the standard curve
A standard curve for determining the abundance of G. smaydae was obtained using DNA extracted from a dense G. smaydae GSSH1005 culture. DNA was extracted from an exponential growth phase culture of G. smaydae, targeting 100,000 cells in the final elution volume of 100 μL using an AccuPrep Genomic DNA extraction kit (Bioneer) according to the manufacturer’s instructions. The extracted DNA was then serially diluted by adding predetermined volumes of DDW to 1.5-mL tubes to ultimately prepare six different DNA concentrations, targeting 100, 10, 1, 0.1, 0.01, and 0.001% DNA concentrations of the originally extracted DNA. The extracted and diluted DNA samples were then stored at −20°C in a freezer and qPCR amplification was conducted within a day. qPCR was used to construct the standard curve using the aforementioned methods and conditions detailed in the specificity analysis section.
Quantification of the abundance of Gymnodinium smaydae
For the determination of the distribution of G. smaydae in Korean waters, qPCR analysis was performed. First, cells in 50–300 mL of each water sample collected from each station at each time interval were collected by filtering through a 25-mm GF/C filter (Whatman Inc., Clifton, NJ, USA). Then, the filter was loosely rolled, placed into a 1.5-mL tube, and frozen at −20°C until it was transported to the laboratory. Subsequently, the DNA was extracted from cells captured on filters using an AccuPrep Genomic DNA extraction kit (Bioneer) according to the manufacturer’s instructions. Extracted DNA was stored at −20°C until qPCR was performed. In addition, to ensure accuracy of results, DNA from each sample was amplified four times. Samples using DDW as the template were used as non-template controls, samples using only DDW as the template and reaction mix were used as negative controls, and DNA used to construct the standard curve was used as positive and standard controls. Conditions used for qPCR and quantification of the abundance of field samples were similar to those listed in the specificity analysis section.
Data analysis
Statistical analyses were performed using the statistical software package SPSS version 25.0 (IBM Corp., Armonk, NY, USA). Pearson’s correlation analysis was used to determine the one-to-one relationship between the cell abundances of G. smaydae and environmental factors.
RESULTS
Spatial and temporal distributions of Gymnodinium smaydae in Korean waters
After developing specific primers and probe for G. smaydae (Table 1), their specificity was tested using five related species and four prey species (Table 2). Primers and probe responded to G. smaydae but not to other species (Fig. 2).
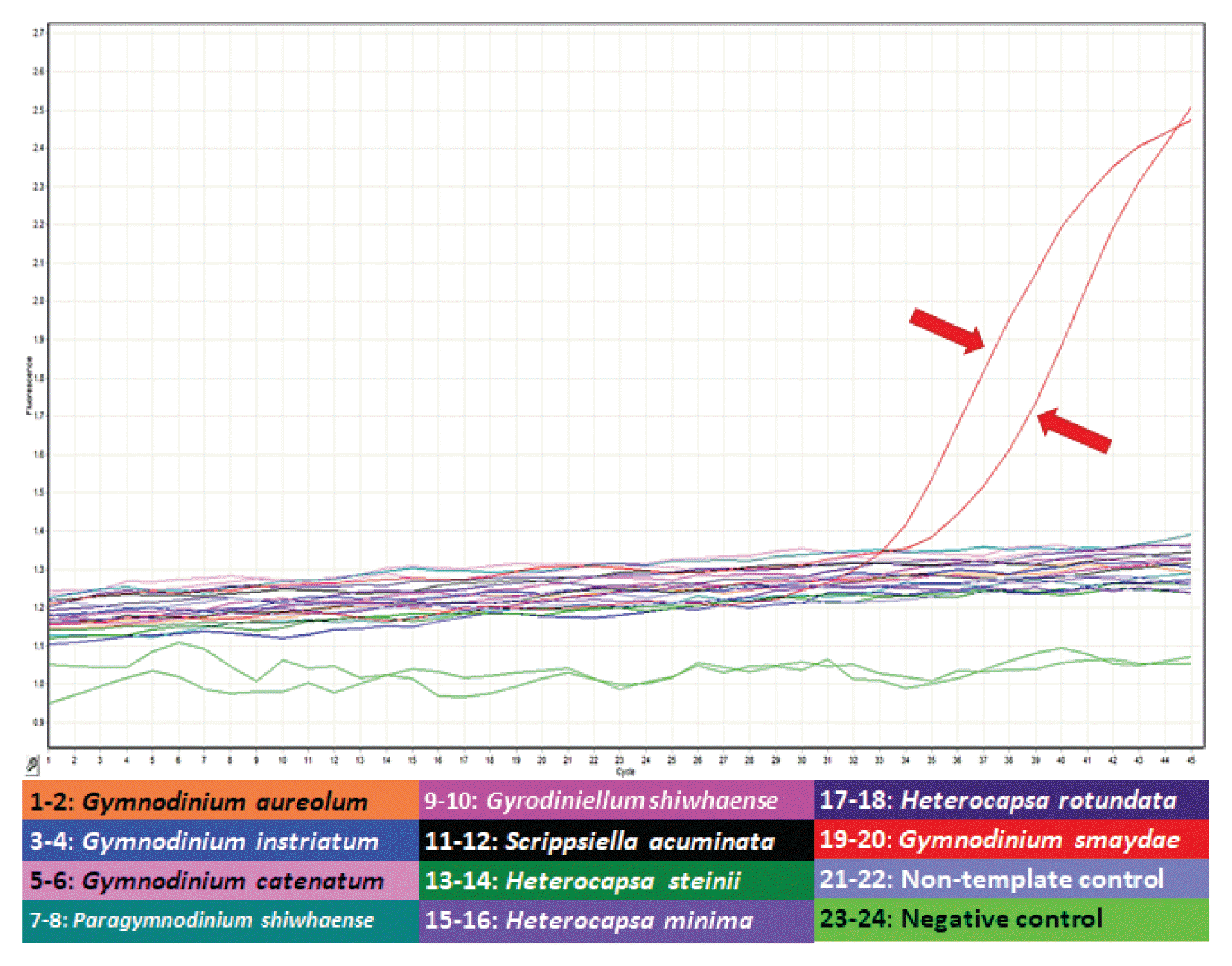
Specificity analysis of specific primers and probe for Gymnodinium smaydae. Amplification plot generated using assays run as a duplicate of samples from four Gymnodinium species (Gymnodinium aureolum, G. catenatum, G. instriatum, and G. smaydae), four prey species of G. smaydae (Heterocapsa rotundata, H. steinii, H. minima, and Scrippsiella acuminata), two dinoflagellates that are morphologically similar to G. smaydae (Paragymnodinium shiwhaense and Gyrodiniellum shiwhaense), non-template controls, and negative controls. Amplification plots obtained from each sample read the increase in fluorescence intensity as the generated polymerase chain reaction product. Red arrows indicate the amplification signal of G. smaydae.
Cells of G. smaydae (≥0.1 cells mL−1) were detected at 23 of 28 sampling stations during the study period (Fig. 1). Cells were not detected in the waters off Ulsan (East Sea), Kwangyang, Goheung (South Sea), Gosan and Wimi (Jeju Island). The highest abundance of G. smaydae (18.5 cells mL−1) was found in Jinhae Bay (South Sea), while the second highest abundance (10.6 cells mL−1) was in the waters off Busan (Table 3, Fig. 3).
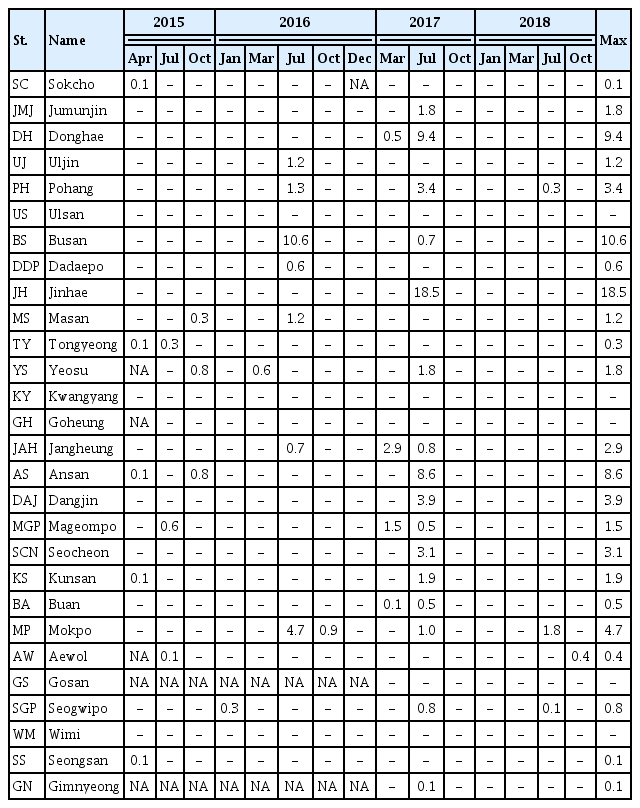
The abundance (cells mL−1) of Gymnodinium smaydae at each station from April 2015 to October 2018, quantified using qPCR
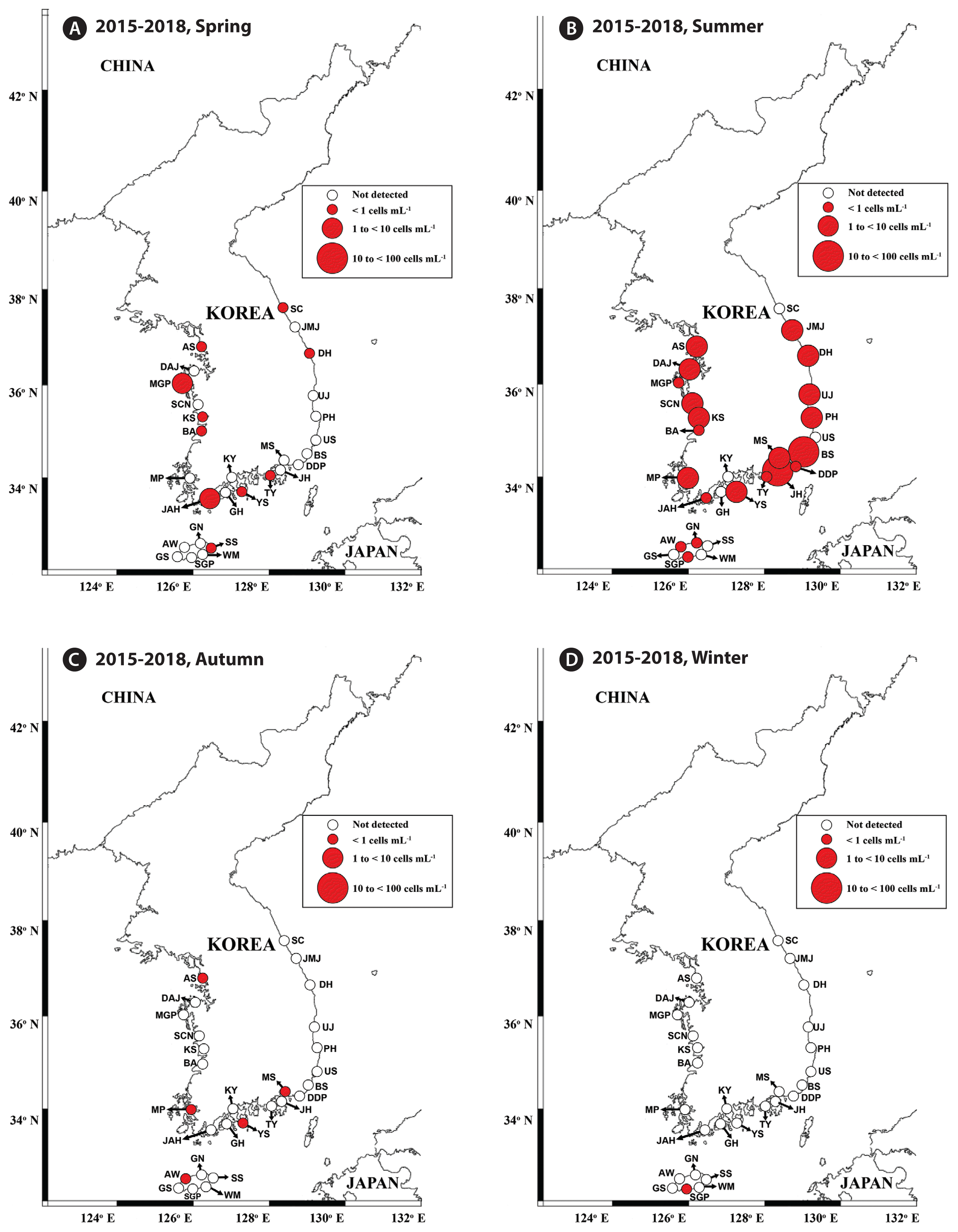
Map of the sampling stations indicating the presence and absence of Gymnodinium smaydae in spring (March or April) (A), summer (July) (B), autumn (October) (C), and winter (December or January) (D) from 2015 to 2018. Red closed circles indicate the stations at which G. smaydae cells were detected, whereas black open circles indicate the stations at which G. smaydae cells were not detected. The size of the red closed circles indicates the abundance of G. smaydae cells. SC, Sokcho; JMJ, Jumunjin; DH, Donghae; UJ, Uljin; PH, Pohang; US, Ulsan; BS, Busan; DDP, Dadaepo; MS, Masan; JH, Jinhae; TY, Tongyeong; YS, Yeosu; KY, Kwangyang; GH, Goheung; JAH, Jangheung; AS, Ansan; DAJ, Dangjin; MGP, Mageompo; SCN, Seocheon; KS, Kunsan; BA, Buan; MP, Mokpo; AW, Aewol; GS, Gosan; SGP, Seogwipo; WM, Wimi; SS, Seongsan; GN, Gimnyeong.
Cells of G. smaydae were detected at 21 stations in summer, 10 stations in spring, and 5 stations in autumn, but detected at only one station (Seogwipo) in winter (Table 3, Fig. 3). Seogwipo is located on Jeju Island. The highest abundance of G. smaydae was found in July 2017 and second highest abundance was in July 2016 (Table 3).
Hydrographic properties during the study period
During the study period, G. smaydae was present in Korean coastal waters when water temperatures were 7.6–28.0°C, salinities were 9.6–34.1, concentrations of NO3 were not detectable (ND)-106.0 μM, and concentrations of PO4 were ND-3.4 μM (Table 4, Fig. 4). Furthermore, G. smaydae was present when the concentrations of SiO2 were ND-448.4 μM and those of DO were 1.7–13.1 mg L−1 (Table 4).
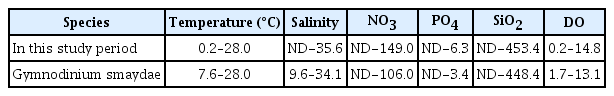
Ranges of the measures of the abiotic environmental factors during the study period and when Gymnodinium smaydae was detected in Korean coastal waters from April 2015 to October 2018
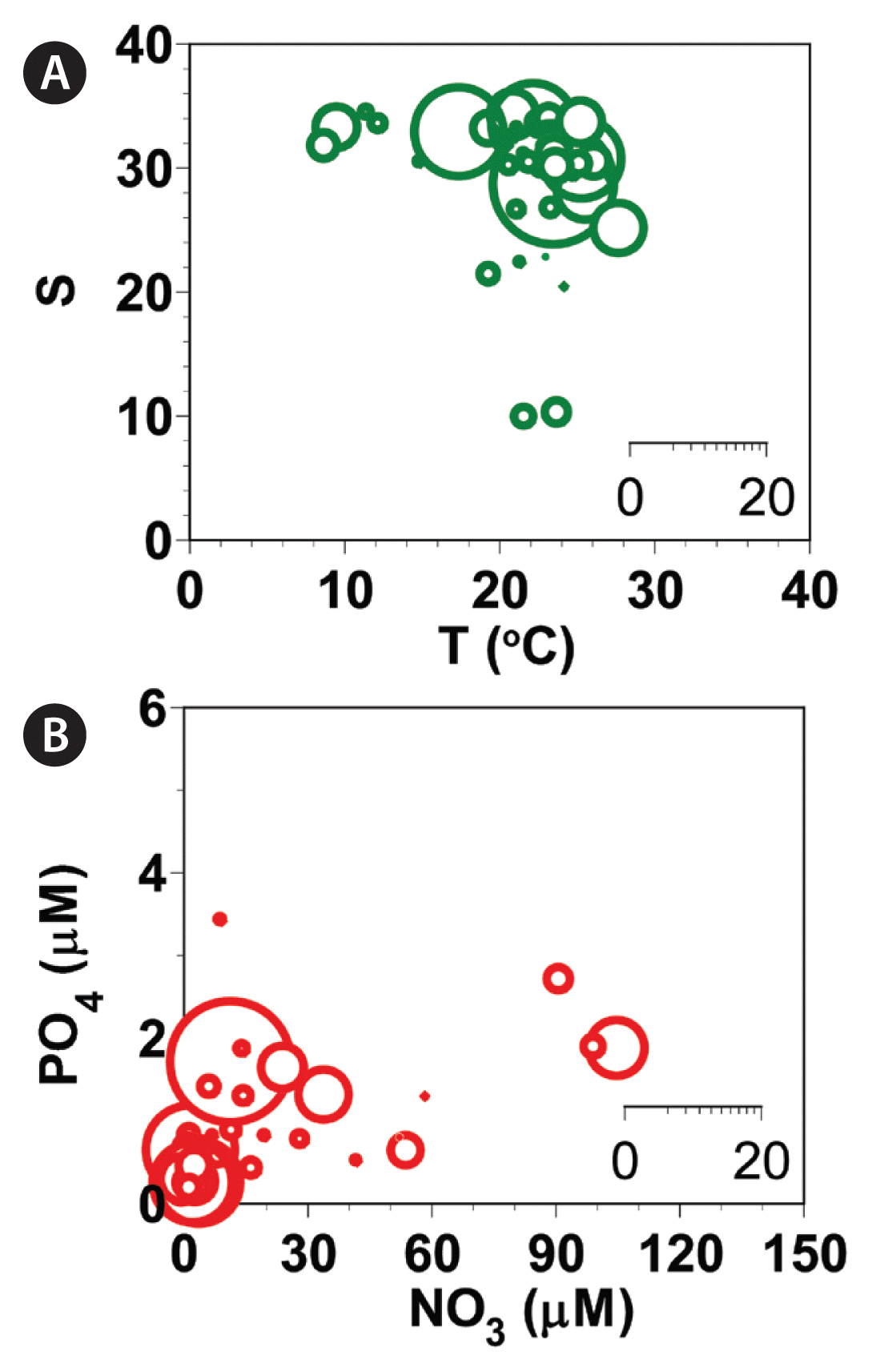
Abundances (cells mL−1) of Gymnodinium smaydae as a function of water temperature (T, °C) and salinity (S) at all stations from 2015–2018 (A) and those of G. smaydae cells as a function of NO3 and PO4 concentrations (B). Scale of circles in the inset is the abundance of G. smaydae cells (cells mL−1).
The highest abundance of G. smaydae was found when the water temperature was 23.8°C, salinity was 28.31, concentration of NO3 was 12.5 μM, concentration of PO4 was 1.65 μM, concentration of SiO2 was 41.0 μM, and concentration of DO was 10.83 mg L−1.
Relationships between the abundance of Gymnodinium smaydae and environmental factors
The Pearson’s correlation analysis showed that the abundance of G. smaydae was significantly and positively correlated with the Chl-a concentration (p < 0.001, r = 0.427) (Table 5). Furthermore, the abundance of G. smaydae was significantly and positively correlated with water temperature (p = 0.002, r = 0.156) (Table 5). However, there were no significant correlations between the abundance of G. smaydae and water salinity, concentrations of nutrients, or DO concentration (Table 5).
DISCUSSION
Cells of G. smaydae have been found in Korean waters ever since this species was described as a new species in 2014 (Kang et al. 2014). Establishing a culture of this species is not easy because it can grow on only a few species (Lee et al. 2014a, You et al. 2020). Furthermore, this dinoflagellate is very small and not easily distinguishable from other small species, such as other Gymnodinium and Paragymnodinium spp., and Gyrodiniellum shiwhaense (Fig. 5). Due to these difficulties, it may be difficult to find this species in the waters of other countries. This study provides the sequences of the specific primers and probe for G. smaydae; thus, the presence of this species can be widely determined.
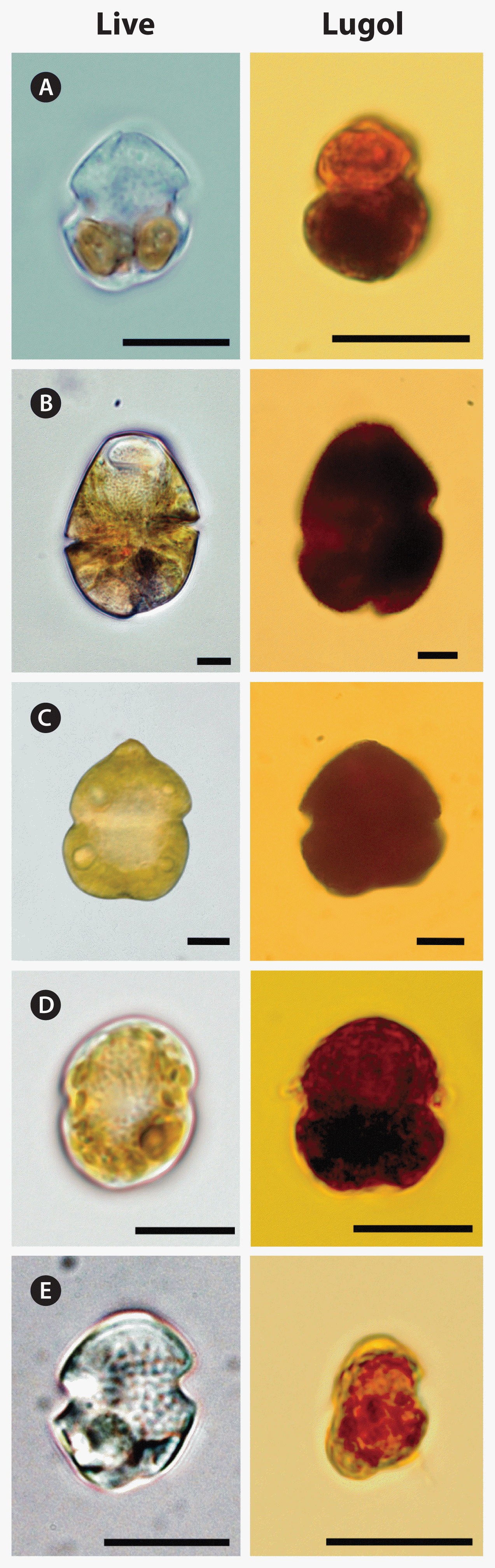
Light micrographs of Gymnodinium smaydae and other dinoflagellate species with similar morphologies. Living cells (left) and cells fixed in 4% Lugol’s solution (right). (A) G. smaydae, (B) Gymnodinium instriatum, (C) Gymnodinium catenatum, (D) Paragymnodinium shiwhaense, and (E) Gyrodiniellum shiwhaense. Scale bars represent: A–E, 10 μm.
The growth and ingestion rates of G. smaydae on H. rotundata and H. steinii in the laboratory have been previously reported (Lee et al. 2014a). Furthermore, the growth and ingestion rates of the heterotrophic dinoflagellate O. marina on G. smaydae have also been reported (Jeong et al. 2018). However, understanding the population dynamics of G. smaydae and its roles in the marine planktonic community was difficult owing to the lack of abundance of G. smaydae in the field. This study provided a range of its abundance and reported the highest abundance of G. smaydae at sea, in addition to providing insights into the effects of environmental factors on the population dynamics of this species.
This study clearly showed that G. smaydae had a wide spatial distribution but a strong seasonality in Korean waters. Furthermore, during the study period, the abundance of G. smaydae was affected by Chl-a concentration, suggesting that the growth or survival of G. smaydae may be affected by prey availability. Heterocapsa minima is known to be present in 26 of the 28 evaluated stations in Korean waters (Lee et al. 2019) and S. acuminata also has a wide distribution in Korean waters (Kim et al. 2019). In addition, red tides caused by H. steinii (= H. triquetra) or Heterocapsa sp. have been observed in Korea (National Institute of Fisheries Science 2020, National Institute of Biological Resources 2020). Therefore, high prey availability may be partially responsible for the wide distribution of G. smaydae in Korean coastal waters.
During the study period, the highest abundance of G. smaydae was 18.5 cells mL−1, and no red tide caused by this species was found. The maximum growth rate of G. smaydae on H. rotundata is 2.23 d−1 (i.e., dividing 3 times per day) (Lee et al. 2014a). The Chl-a concentration at the water where the highest abundance of G. smaydae was found was 127 ng Chl-a mL−1. If this Chl-a belonged to H. rotundata (1.1 pg Chl-a per cell), the calculated abundance of H. rotundata would be 115,000 cells mL−1. When the equation for the mixotrophic growth rate of G. smaydae on H. rotundata in Lee et al. (2014a) was used, the calculated growth rate of G. smaydae at this Chl-a concentration was 2.22 d−1. Thus, phytoplankton other than Heterocapsa spp. and S. acuminata might be major contributors to Chl-a concentration and might not support the growth of G. smaydae.
Gymnodinium smaydae cells were present at most stations in summer but were present at only one station in winter. You et al. (2020) reported that the maximum mixotrophic growth rate of G. smaydae on H. rotundata was positive at 10–32°C, but negative at ≤8°C. During this study period, G. smaydae was present in Korean coastal waters at 7.6–28.0°C, although the range of water temperatures at all stations was 0.2–28.0°C. Therefore, G. smaydae is not likely to survive in water temperatures of <7–8°C. In winter, G. smaydae was only found in waters at Seogwipo, where the water temperature was 15.1°C. Thus, a water temperature >10°C in winter may have allowed G. smaydae to survive in the water at Seogwipo. You et al. (2020) also reported that the maximum mixotrophic growth rate was observed at 25°C. During this study period, the water temperature at which the highest abundance of G. smaydae was found was 23.8°C. Therefore, results of the laboratory experiments were consistent with those of the field observations, indicating that G. smaydae preferred temperatures of approximately 25°C.
Water temperature in Korean coastal waters in summer sometimes exceeds 30°C (Ha et al. 1998, Kim and Park 2018, Seo et al. 2020). Global warming is expected to elevate water temperatures of Korean waters in summer (Kim and Park 2018, Seo et al. 2020). Gymnodinium smaydae cells can grow at 32°C, while many other dinoflagellates have been reported to die at this temperature (Hand et al. 1965, Kamykowski and McCollum 1986, Morton et al. 1992, Nitschke et al. 2015, Jang et al. 2018, Lim et al. 2019b, Ok et al. 2019, You et al. 2020). Therefore, G. smaydae may have an advantage over other dinoflagellates during hot summers. The combination of the data from laboratory experiments and field observations can help improve our understanding of the population dynamics of G. smaydae and its roles in the marine planktonic community.
ACKNOWLEDGEMENTS
We are grateful to An Suk Lim, Se Hyeon Jang, Sang Ah Park, Se Hee Eom, Jae Yeon Park, and Jung Rye Rho for technical support. This work was supported by the useful dinoflagellate program of Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) and the National Research Foundation (NRF) funded by the Ministry of Science and ICT (NRF-2017R1E1A1A01074419) award to HJJ.