ABSTRACTOomycetes are ubiquitous heterotrophs of considerable economic and ecological importance. Lately their diversity in marine environments has been shown to be greatly underappreciated and many lineages of intracellular holocarpic parasites, infecting micro- and macro-algae, remain to be fully described taxonomically. Among them, pathogens of marine red algae have been studied extensively as they infect important seaweed crops. Throughout the 20th century, most intracellular, holocarpic biotrophic oomycetes that infect red algae have been assigned to the genus Olpidiopsis Cornu. However, 18S rRNA sequencing of Olpidiopsis saprolegniae, the species considered the generitype for Olpidiopsis, suggests that this genus is not closely related to the marine pathogens and that the latter requires a nomenclatural update. Here, we compile and reanalyze all recently published 18S rRNA sequence data for marine holocarpic oomycetes, with a particular focus on holocarpic pathogens of red algae. Their taxonomy has been revised twice over the past four years, with suggestions to transfer them first into the genus Pontisma and then Sirolpidium, and into a monogeneric order, Pontismatales. We show however, that previously published topologies and the proposed taxa Pontisma, Sirolpidium, and Pontismatales are unsupported. We highlight that name changes that are unfounded and premature create confusion in interested parties, especially concerning pathogens of marine red algae that infect important seaweed crops. We thus propose that the names of these holocarpic biotrophic parasites of red algae are retained temporarily, until a supported topology is produced with more genetic markers to enable the circumscription of species and higher-level taxa.
INTRODUCTIONOver the past five years, an unrecognized diversity of aquatic oomycetes has been unveiled by many independent studies using field observations, metabarcoding, single-cell isolation of phytoplankton, and even time-consuming laboratory cultures. To date, these organisms appear to form a hugely diverse group of intracellular holocarpic parasites that infect marine and freshwater algae: green algae (Chlorophyta), red algae (Rhodophyta), brown algae (Phaeophyceae), and diatoms (Bacillariophyceae). They have been suggested to play potentially important roles in controlling phytoplankton dynamics (Garvetto et al. 2018) and represent a known or potential threat for commercially cultivated algae (Badis et al. 2019, 2020). The severe economic costs of these pathogens have increased with the spread and intensification of seaweed (esp. red algal) aquaculture over the last decades (Kim et al. 2014, Wen et al. 2023).
These intracellular, obligately parasitic oomycetes infecting algae have started being described in the second half of the 19th century (Fig. 1), with the delineation of taxa based on a combination of morphological characters (e.g., flagellation of zoospores) and life cycle (e.g., morphogenesis of sporangia). These early studies also suggested that host specificity could have taxonomic utility, yet this needed further study (Sparrow 1960). Later studies on laboratory cultures indicated that parasites had greater host diversity than originally assumed from field observations (Sekimoto et al. 2008, 2009, Fletcher et al. 2015, Klochkova et al. 2016, 2017). Until the first molecular sequences became available at the turn of this century, the phylogenetic position of these oomycetes has remained all but uncertain, and some have long been misidentified as chytrids or even hyphochytrids (Gachon et al. 2017).
Olpidiopsis was originally described as an endobiotic parasite of various oomycetes and freshwater and marine algae (Cornu 1872, Sparrow 1960). The type species is O. saprolegniae (A. Braun) Cornu, a parasite of Saprolegnia spp. Recently, a parasite infecting Saprolegnia parasitica was isolated and barcoded, this host species matched the original host (Saprolegnia spp.) (Braun 1855) and its pathogen matched O. saprolegniae (Cornu 1872), leading to its designation as an epitype (Buaya et al. 2019). Several holocarpic pathogens of red algae have historically also been placed in the genus Olpidiopsis, due to their morphological similarity with holocarpic pathogens of oomycetes. They share a usually single spherical holocarpic zoosporangium, with one or several evanescent discharge tubes (Sparrow 1960), yet appear to be phylogenetically distant from the epitype of O. saprolegniae (Buaya et al. 2019). These species of red algal parasites for which 18S rRNA data is available, widely known as Olpidiopsis bostrychiae, O. feldmanni, O. heterosiphoniae, O. muelleri, O. palmariae, O. porphyrae, and O. pyropiae, therefore cannot be included in this genus any more (Buaya et al. 2019, 2023a, Buaya and Thines 2020).
Another intracellular pathogen of red algae is Pontisma lagenidioides H. E. Petersen (1905), which forms two types of sporangia: simple “olpidiod” sporangia, and sporangia stemming from fragmenting thalli (Petersen 1905, Sparrow 1960). Recently, a parasite of an alga identified as Ceramium rubrum which had a “thallus […] usually composed of a series of somewhat irregularly cylindrical, sausage-like segments” was sequenced and used to epitypify the genus Pontisma (as P. lagenidiodes). Following an earlier speculation by Dick (2001), these authors then transferred all holocarpic parasites of red algae into the genus Pontisma though this clade was unsupported in their phylogenetic analyses (Buaya et al. 2019, Buaya and Thines 2020).
Sirolpidium H.E. Petersen was originally erected alongside Pontisma to account for the ability of its type, S. bryopsidis to differentiate two types of thalli (olpidioid and fragmenting) within its green algal host Bryopsis plumosa (Petersen 1905, Sparrow 1960). The morphogenetic similarities between Pontisma and Sirolpidium had long been recognized and led to the suggestion of merging both genera (Karling 1942). Recently, a pathogen of the green filamentous Ulotrichales, Capsosiphon fulvescens, tentatively likened to Sirolpidium bryopsidis, despite its apparent inability to infect Bryopsis plumosa and different geographic origin, was found to have a partial 18S sequence identical with Pontisma lagenidioides (Buaya et al. 2021). Finally, a novel pathogen of a second green filamentous Ulotrichales, Urospora neglecta, which only showed olpidioid thalli and had a 18S sequence that was sister, though with only moderate bootstrap support, to the red algal parasite Olpidiopsis porphyrae. Having realized that Karling retained Sirolpidium over Pontisma, these authors now propose to assign all holocarpic, pathogens of red and green algae mentioned above to the genus Sirolpidium, within the monogeneric order Pontismatales (Buaya et al. 2023b). As defined in the latter paper, this group also encompasses pathogens of diatoms and brown algae (Garvetto et al. 2018, Buaya and Thines 2022), as well as environmental sequences from uncharacterized organisms.
This body of work has led to repeated name changes to some important red algal pathogens twice over the past four years (Buaya et al. 2019, 2023b), though it was obvious from all these papers that this new circumscription of Pontisma (or lately, Sirolpidium) and the order Pontismatales was unsupported in any analyses (e.g., Badis et al. 2019, Buaya et al. 2019, 2021, 2023b, Buaya and Thines 2020). We decided to reinvestigate the phylogeny of this group based on the same data set of publicly available small subunit ribosomal DNA (SSU rDNA). Our results confirm that the taxa Pontisma, Sirolpidium, and Pontismatales are not supported. We discuss the negative consequences of premature nomenclatural changes to stakeholders and make a recommendation to conserve incorrect, yet widely understood, original names of red algal pathogens until a supported phylogeny can be obtained and adequate nomenclatural changes made.
MATERIALS AND METHODSThe nuclear encoded small subunit ribosomal 18S RNA gene (SSU) dataset was copied Buaya and Thines (2020) with added all recent sequences of holocarpic biotrophic algal parasites (e.g., Buaya et al. 2023b) from GenBank. Several alignment methods were used, that all employ different algorithms and penalty scores: (1) Pagan using the Was@bi webserver (http://was.bi/) and default parameters (Löytynoja 2021); (2) PRANK alignment using the stand alone package also incorporating the default parameters (Löytynoja and Goldman 2010); (3) T-Coffee for multiple DNA sequence alignment (M-Coffee) (Notredame et al. 2000) used the webserver (https://tcoffee.crg.eu/apps/tcoffee/) (Di Tommaso et al. 2011); (4) the SINA aligner (Preusse et al. 2012) on the SILVA website (https://www.arb-silva.de) (Quast et al. 2012); and finally, (5) we used MAFFT with the alignment strategy Q-INS-I, on the online server (https://mafft.cbrc.jp/alignment/server/) (Katoh and Standley 2013, Katoh et al. 2019).
Maximum-likelihood analyses were implemented using IQ-TREE 2 (Minh et al. 2020) and Bayesian inference analysis with MrBayes v. 3.2 (Ronquist et al. 2012). Model selection (Kalyaanamoorthy et al. 2017) implemented in IQ-TREE. Non-parametric bootstrapping (500 replicates) (Felsenstein 1985) was used to determine branch for the main tree (MAFFT aligned); 100 replicates for supplementary trees (and Bayesian analysis omitted for these trees).
Bayesian inference used variable rates and six rate categories. Two parallel runs of Markov chain Monte Carlo were performed for 3,000,000 generations, sampling every 1,000 generations. Estimated samples size, split frequencies, and stationarity were checked after each run. Post-analysis, 10% of generations were removed as a burn-in and posterior probabilities visualized in Figtree v1.1.4 (Rambaut 2009) and Canvas X Draw (Canvas GFX, Inc., Boston, MA, USA) were used to manipulate trees (e.g., collapse branches) for presentation.
The outgroups for all analyses were as in Buaya and Thines (2020): Hyphochytrium catenoides (AF163294, X80344), Developayella elegans (U37107), and an uncultured eukaryote (AB695482).
RESULTS AND DISCUSSIONTo maximize the phylogenetic signal, we retained the maximum number of base pairs in the alignment so that after trimming, less than 20% of the partial SSU sequences were below 850 bp long. The different algorithms produced alignments of various lengths, with various numbers of variable (and parsimony-informative) sites and trees with different maximum-likelihoods (Table 1). All our analyses, as well as all previously published analyses (e.g., Buaya et al. 2019, 2023b, Buaya and Thines 2020) concur that the four supported clades containing holocarpic red algal parasites do not cluster together (Fig. 2), so that grouping these lineages into any single genus or single order (Fig. 2) is unwarranted. Even with the same data set, and different alignment methods, the relationships between these unsupported clades, as expected, vary (Supplementary Fig. S1).
Our investigation of the partial 18S sequences available for Pontisma lagenidioides and Sirolpidium bryopsidis (MK253530.2 and MW489460.1) further reveals that they are not identical over their entire length (818 bp) though conspecificity between both parasites was inferred because of “no genetic distance” between them (Buaya et al. 2023b). The identifications of the organisms concerned are also questionable: on the one hand, Sirolpidium bryopsidis had never been found before infecting Capsosiphon fulvescens. The latter host was collected in Iceland, whereas the type was described from Frederikshavn (Denmark), so that direct evidence is lacking that Sirolpidium bryopsidis is indeed the organism sequenced in Buaya et al. (2021). On the other hand, the host red alga depicted as the host for the epitype of Pontisma lagenidioides, is incompletely corticated (Buaya et al. 2019). Therefore, it is not C. rubrum as stated by the authors, and is in fact an unidentified Ceramium sp. These two examples illustrate that defining epitypes is only useful, to overcome the limitations of original taxonomic descriptions and to link them with molecular data, if utmost care is taken in the identification of host, choice of sampling location, and subsequent data analysis.
The novel parasite, Sirolpidium litorale, isolated from the green alga Urospora neglecta neither forms a clade with the known SSU sequence for S. bryopsidis (Buaya et al. 2023b, and our Fig. 1), nor seems to present typical features of this genus, such as fragmenting thalli (Petersen 1905, Sparrow 1960).
The absence of monophyly between the clades of red algal pathogens designated Pontisma or Sirolpidium by Buaya and colleagues, is now undisputable. Hence, the proposed circumscription of the genus Pontisma, or Sirolpidium, as a taxon encompassing pathogens of red algae, or for that matter any specific algal host, will have to be revised; instead, with the caveat highlighted above concerning the correct identification of its epitype, we propose that the genus name Pontisma should be restricted to Pontisma lagenidioides. We also reject the proposed synonymy between Pontisma and Sirolpidium for the time being and propose that the latter genus name remains restricted to its type, Sirolpidium bryopsidis, at least until (1) its presumed conspecificity with the parasite infecting Capsosiphon fulvescens is confirmed through the sequencing of endoparasites of Bryospis plumosa; and (2) a suitable epitype can be defined.
The Pontismatales, which has never been supported in any analyses should be disused. It was suggested that it could encompass red algal pathogens (Buaya and Thines 2020) but this was not correct, even in the Buaya and Thines (2020) analysis, where this was highlighted, it included diatom pathogens and subsequently found to also likely encompass brown algal pathogens (Buaya and Thines 2022, Buaya et al. 2023a). Our analyses clearly show that there are currently no groups that could be placed in a monophyletic supported order of biotrophic algal pathogens, as recently suggested (Buaya et al. 2023b). If it were confirmed as a monophyletic group, the concept of this order would require being broadened to accommodate far greater diversity of hosts (including brown algae and green algae), morphologies, and spore morphogenesis than is currently the case.
Over the last four years, two novel generic names, and the accompanying nomenclatural changes, Pontisma and Sirolpidium, have been proposed by Buaya et al. to designate species of parasites of marine red algae: Olpidiopsis bostrychiae, O. feldmanni, O. heterosiphoniae, O. muelleri, O. palmariae, O. porphyrae, and O. pyropiae. For the reasons stated above, it is hard to see any improvement over the original genus name Olpidiopsis, which at least has the merit of being widely used, especially by applied scientists and seaweed professionals from the industry and government bodies. Even when justifiable from a taxonomic point of view, name changes for ecologically significant and economically important species have deep consequences on society, potentially affecting livelihoods or conservation policies. Consequently, we argue in favor of retaining the names of these taxa until a robustly-supported nomenclatural revision can be performed.
Though we are aware of the challenges hindering the description and classification of holocarpic oomycetes, we feel that novel taxa should not be described without the provision of several congruent markers: indeed, relying on partial 18S sequences is insufficient. While many of these original descriptions are very insightful (e.g., Braun 1855, Sparrow 1960), attaching names to newly collected specimens, often on different hosts and different locations, from the original descriptions that were based on few characters (i.e., general similarity, sexual characters are missing in all red algal holocarpic pathogens) just brings more confusion, and retards naming and describing new taxa. We recommend that the nomenclature of supra-specific taxa should only be emended to produce a natural classification, when the corresponding taxa are precisely circumscribed with robust phylogenetic, morphological, and possibly ultrastructural or ecological criteria. Acquiring phylogenomic evidence on these taxa is most likely to be key to enable such a step.
CONCLUSIONThe full diversity of biotropic oomycete pathogens is just now being explored. Our understanding of their relationships is still hampered by a lack of datasets that resolve their relationships. Premature nomenclatural changes based on unsupported relationships do not aid in furthering research, nor are they useful to interested parties. We propose that algal pathogens named “Olpidiopsis” remain that way until further data can be gathered to resolve relationships and more careful epitypification of old names be performed.
ACKNOWLEDGEMENTSThis work was supported by the management of Marine Fishery Bio-resources Center (2024) funded by the National Marine Biodiversity Institute of Korea (MABIK) and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (NRF-2022R1A2C1091633) and the development of technology for biomaterialization of marine fisheries by-products of Korea institute of Marine Science & Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (KIMST-20220128).
SUPPLEMENTARY MATERIALSSupplementary Fig. S1Maximum-likelihood topology of SSU sequence data of various Oomycota (https://www.e-algae.org). Fig. 1Morphology of the main taxa of endobiotic, holocarpic oomycete pathogens mentioned in the text. (A) The generitype Olpidiopsis saprolegniae infecting Saprolegnia sp., as first illustrated by Cornu (1872, left), and its epitype (right). (B–F) Pathogens of red algae commonly assigned to Olpidiopsis since the second half of the 20th century. (B) O. bostrychiae infecting Bostrychia moritziana. Arrowheads, zoosporanium developing liberation tube; double arrowhead; just infected host cell. (C) O. heterosiphoniae infecting Heterosiphonia pulchra. Arrow, zoosporanium. (D) O. feldmanni infecting Asparagopsis sp. Arrow, liberation tube of the zoosporangium. (E) O. palmariae infecting tetraspores of Palmaria palmata. (F) Olpidiopsis species infecting Porphyra and Pyropia spp. O. muelleri infected Porphyra sp. blade. (G) O. porphyrae infecting P. yezoensis. (H) O. pyropiae infecting P. yezoensis. (I) DAPI staining of O. pyropiae infecting P. yezoensis. (J) Zoosporangium of O. porphyrae developing in a P. yezoensis cell (white arrow); empty sporangium (black arrows). (K) The generitype Pontisma lagenidioides infecting Ceramium rubrum, as first illustrated by Petersen (1905, left), and its epitype (right). (L) The generitype Sirolpidium bryopsidis infecting Bryopsis plumosa as first illustrated by Petersen (1905, left), and the morphologically similar oomycete identified as such infecting Capsosiphon fulvescens (right). (M–O) Other oomycete taxa of endobitiotic, holocarpic parasites of algae with incompletely resolved relationships with the abovementioned pathogens of red algae. (M) Sirolpidium litorale in Urospora neglecta. Red arrows, liberation tubes. (N) Pontisma lauvikense infecting the filamentous brown alga Pylaiella littoralis. (O) An unnamed parasitoid of the diatom Licmophora sp. Scale bars represent: A, C–E, G, I, K, L & O, 50 μm; B, H, J & M, 20 μm; F, 1 cm; N, 100 μm. 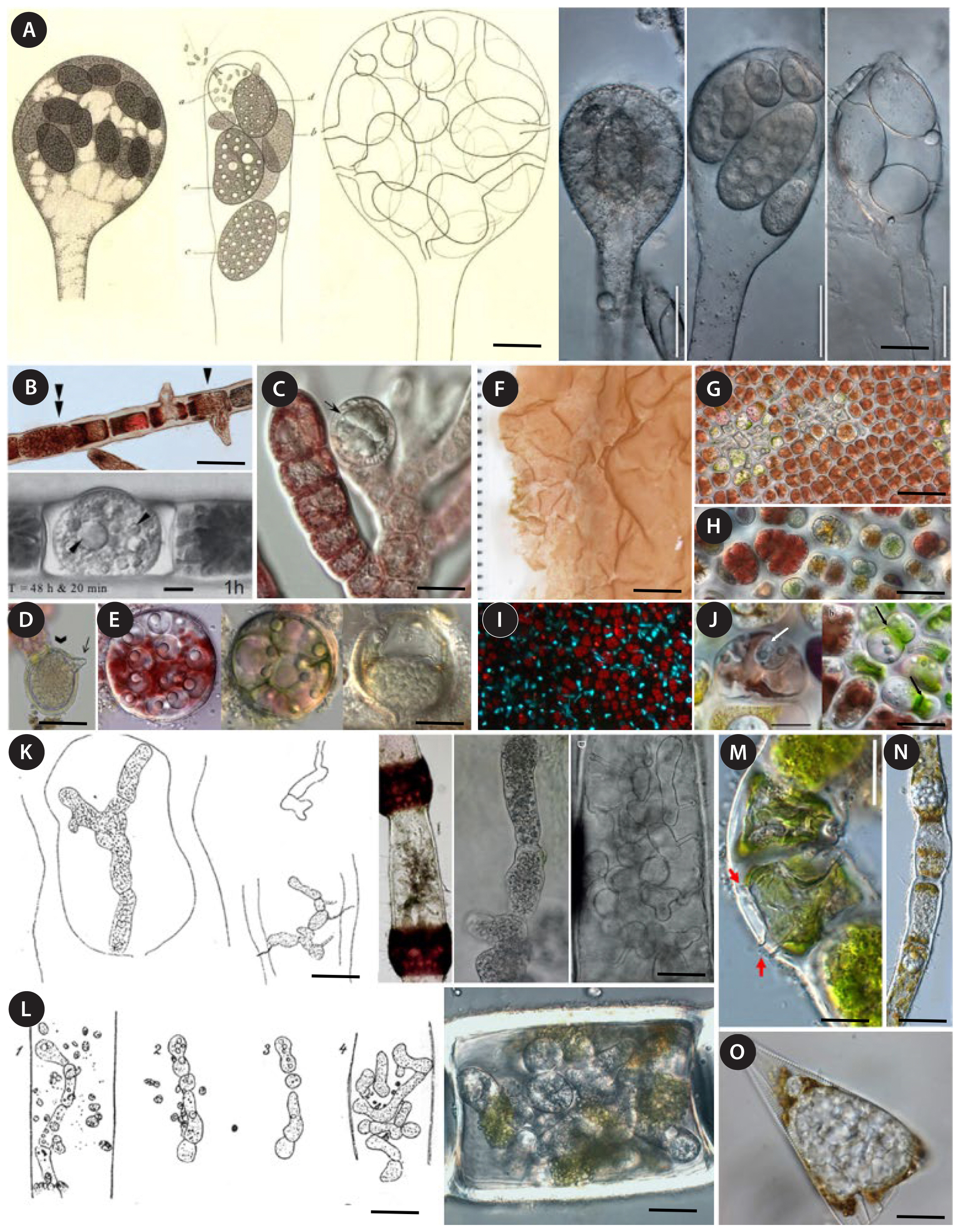
Fig. 2Cartoon of maximum-likelihood topology of small subunit ribosomal 18S RNA gene (SSU) sequence (aligned with MAFFT) data of various Oomycota (following Buaya et al. 2019, 2023b). Lined boxes show specimens placed in the order Pontismatales, or the single genus Pontisma or Sirolpidium at various times. Host of the pathogens shown: red box, red algal parasites; green box, green algal parasites; brown box, brown algal or diatom parasites. Purple box shows the unsupported backbone phylogeny based in these SSU sequences. Arrow indicated the type for Olpidiopsis. Number on branches are non-parametric bootstrap in % and Bayesian posterior probabilities (values <70% and <0.90 removed). 
Table 1Alignments and maximum-likelihood scores for SSU of oomycete taxa REFERENCESBadis, Y., Klochkova, T. A., Brakel, J. & et al 2020. Hidden diversity in the oomycete genus Olpidiopsis is a potential hazard to red algal cultivation and conservation worldwide. Eur. J. Phycol. 55:162–171.
doi.org/10.1080/09670262.2019.1664769
Badis, Y., Klochkova, T. A., Strittmatter, M. & et al 2019. Novel species of the oomycete Olpidiopsis potentially threaten European red algal cultivation. J. Appl. Phycol. 31:1239–1250.
doi.org/10.1007/s10811-018-1641-9
Braun, A. 1855. Ueber Chytridium eine Gattung einzelner Schmarotzerge-wachse auf Algen und Infusorien. Monatsb. Königl. Preuss. Akad. Wiss. Berlin. 1856:21–83.
Buaya, A. T., Ploch, S., Inaba, S. & Thines, M. 2019. Holocarpic oomycete parasitoids of red algae are not Olpidiopsis. Fungal Syst. Evol. 4:21–31.
doi.org/10.3114/fuse.2019.04.03
Buaya, A. T., Scholz, B. & Thines, M. 2021.
Sirolpidium bryopsidis, a parasite of green algae, is probably conspecific with Pontisma lagenidioides, a parasite of red algae. Fungal Syst. Evol. 7:223–231.
doi.org/10.3114/fuse.2021.07.11
Buaya, A. T. & Thines, M. 2020. An overview on the biology and phylogeny of the early-diverging oomycetes. Philipp. J. Syst. Biol. 14:1–20.
doi.org/10.26757/pjsb2020a14004
Buaya, A. & Thines, M. 2022.
Miracula blauvikensis: a new species of Miracula from Iceland, and report of a co-cultivation system for studying oomycete-diatom interactions. Fungal Syst. Evol. 10:169–175.
doi.org/10.3114/fuse.2022.10.07
Buaya, A., Tsai, I. & Thines, M. 2023a.
Pontisma blauvikense sp. nov. the first member of the early-diverging oomycete genus Pontisma parasitizing brown algae. J. Eukaryot. Microbiol. 70:e12957
doi.org/10.1111/jeu.12957
Buaya, A. T., Tsai, I., Klochkova, T. A. & Thines, M. 2023b. Introducing a new pathosystem for marine pathogens: the green alga Urospora neglecta and its pathogen Sirolpidium litorale sp. nov. Mycol. Prog. 22:86
doi.org/10.1007/s11557-023-01938-w
Cornu, M. 1872. Monographie des Saprolegniees, etude physiologique et systematique. Ann. Sci. Nat. Bot. 15:1–198.
Dick, M. W. 2001. Straminipilous fungi: systematics of the Peronosporomycetes including accounts of the marine straminipilous protists, the plasmodiophorids and similar organisms. Kluwer Academic Publishers, Dordrecht, 670 pp.
Di Tommaso, P., Moretti, S., Xenarios, I. & et al 2011. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39(Suppl 2):W13–W17.
doi.org/10.1093/nar/gkr245
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39:783–791.
doi.org/10.2307/2408678
Fletcher, K., Žuljević, A., Tsirigoti, A. & et al 2015. New record and phylogenetic affinities of the oomycete Olpidiopsis feldmanni infecting Asparagopsis sp. (Rhodophyta). Dis. Aquat. Org. 117:45–57.
doi.org/10.3354/dao02930
Gachon, C. M. M., Strittmatter, M., Badis, Y., Fletcher, K. I., Van West, P. & Müller, D. G. 2017. Pathogens of brown algae: culture studies of Anisolpidium ectocarpii and A. rosenvingei reveal that the Anisolpidiales are uniflagellated oomycetes. Eur. J. Phycol. 52:133–148.
doi.org/10.1080/09670262.2016.1252857
Garvetto, A., Nézan, E., Badis, Y. & et al 2018. Novel widespread marine oomycetes parasitising diatoms, including the toxic genus Pseudo-nitzschia: genetic, morphological, and ecological characterisation. Front. Microbiol. 9:2918
doi.org/10.3389/fmicb.2018.02918
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 14:587–589.
doi.org/10.1038/nmeth.4285
Karling, J. S. 1942. The simple holocarpic biflagellate Phycomycetes including a complete host index and bibliography. The Author, New York, 123 pp.
Katoh, K., Rozewicki, J. & Yamada, K. D. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20:1160–1166.
doi.org/10.1093/bib/bbx108
Katoh, K. & Standley, D. M. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30:772–780.
doi.org/10.1093/molbev/mst010
Kim, G. H., Moon, K.-H., Kim, J.-Y., Shim, J. & Klochkova, T. A. 2014. A revaluation of algal diseases in Korean Pyropia (Porphyra) sea farms and their economic impact. Algae. 29:249–265.
doi.org/10.4490/algae.2014.29.4.249
Klochkova, T. A., Jung, S. & Kim, G. H. 2016. Host range and salinity tolerance of Pythium porphyrae may indicate its terrestrial origin. J. Appl. Phycol. 29:371–379.
doi.org/10.1007/s10811-016-0947-8
Klochkova, T. A., Kwak, M. S. & Kim, G. H. 2017. A new endoparasite Olpidiopsis heterosiphoniae sp. nov. that infects red algae in Korea. Algal Res. 28:264–269.
doi.org/10.1016/j.algal.2017.09.019
Löytynoja, A. 2021. Phylogeny-aware alignment with PRANK and PAGAN. In : Katoh K., editor Multiple Sequence Alignment: Methods and Protocols. Humana, New York, 17–37.
Löytynoja, A. & Goldman, N. 2010. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics. 11:579
doi.org/10.1186/1471-2105-11-579
Minh, B. Q., Schmidt, H. A., Chernomor, O. & et al 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37:1530–1534.
doi.org/10.1093/molbev/msaa015
Notredame, C., Higgins, D. G. & Heringa, J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–217.
doi.org/10.1006/jmbi.2000.4042
Petersen, H. E. 1905. Contributions à la connaissance des phycomycètes marins (Chytridinae Fischer). Overs. Kongel. Danske Vidensk. Selsk. Forh. 5:439–488.
Pruesse, E., Peplies, J. & Glöckner, F. O. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 28:1823–1829.
doi.org/10.1093/bioinformatics/bts252
Quast, C., Pruesse, E., Yilmaz, P. & et al 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596.
doi.org/10.1093/nar/gks1219
Rambaut, A. 2009. FigTree. Tree figure drawing tool, Available from: http://tree.bio.ed.ac.uk/software/figtree/. Accessed Jan 31, 2024
Ronquist, F., Teslenko, M., van der Mark, P. & et al 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542.
doi.org/10.1093/sysbio/sys029
Sekimoto, S., Klochkova, T. A., West, J. A., Beakes, G. W. & Honda, D. 2009.
Olpidiopsis bostrychiae sp. nov.: an endoparasitic oomycete that infects Bostrychia and other red algae (Rhodophyta). Phycologia. 48:460–472.
doi.org/10.2216/08-11.1
Sekimoto, S., Yokoo, K., Kawamura, Y. & Honda, D. 2008. Taxonomy, molecular phylogeny, and ultrastructural morphology of Olpidiopsis porphyrae sp. nov. (Oomycetes, Straminipiles), a unicellular obligate endoparasite of Bangia and Porphyra spp. (Bangiales, Rhodophyta). Mycol. Res. 112:361–374.
doi.org/10.1016/j.mycres.2007.11.002
Sparrow, F. K. 1960. Aquatic Phycomycetes. University of Michigan Press, Ann Arbor, MI, 1187.
Wen, X., Zuccarello, G. C., Klochkova, T. A. & Kim, G. H. 2023. Oomycete pathogens, red algal defense mechanisms and control measures. Algae. 38:203–215.
doi.org/10.4490/algae.2023.38.12.13
|
|
||||||||||||||||||||||||||||||||||||||||