Abbreviations
ACE
angiotensin-converting enzyme
BSL3
biosafety level 3
CHARMM
Chemistry at Harvard Macromolecular Mechanics
FDA
U.S. Food and Drug Administration
KCDC
Korea Centers for Disease Control and Prevention
MD
molecular dynamics
MERS-CoV
Middle East respiratory syndrome coronavirus
Mpro
main protease
PDB
Protein Data Bank
RdRp
RNA-dependent RNA polymerase
RMSD
root-mean-square deviation
ROS
reactive oxygen species
SARS-CoV
severe acute respiratory syndrome coronavirus
WHO
World Health Organization
INTRODUCTION
Human coronavirus diseases have repeatedly emerged since the beginning of the 21th century; for instance, the emergence of severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002–2003, human coronavirus NetherLand 63 in 2004, HCoV-Hong Kong University 1 in 2005, Middle East respiratory syndrome coronavirus (MERS-CoV) in 2011, and SARS-CoV-2 in 2019 (Liu et al. 2021). Particularly, SARS-CoV-2 caused a global pandemic with substantial morbidity and mortality—approximately 7,000,000 deaths were confirmed as of May 2023 (World Health Organization [WHO], updated May 2023).
Although WHO no longer considers SARS-CoV-2 to be a Public Health Emergency of International Concern, viral infections still remain a persistent public health issue. Many studies concerned that the viral disease emergence such as re-emergence of SARS-CoV and MERS-CoV, and even the emergence of new viruses, is expected to accelerate and repeat (Totura and Bavari 2019, da Silva Antonio et al. 2020, Meganck and Baric 2021); these risks suggest the need to continue research on antiviral drug discovery against various viruses, including SARS-CoV-2.
SARS-CoV-2 spike protein attaches to angiotensin-converting enzyme (ACE) 2 cellular receptor, and the virus enters the host cells and releases the genomic RNA. Immediately, the genomic RNA is translated to open reading frame 1a and 1b, and the resulting polyproteins pp1a and pp1ab are processed into 16 nonstructural proteins such as the various enzymes and transcription factors. These nonstructural proteins form the viral replication and transcription complexes and operate the enzymatic functions involved in RNA synthesis, RNA proofreading, and RNA modification. Also, translated structural proteins translocate into endoplasmic reticulum membranes, and the virion matures and finally secretes from the infected cell by exocytosis (Panda et al. 2020, Pandey et al. 2020, V’kovski et al. 2021, Kang et al. 2023).
Natural products include a plethora of compounds that can be used for the identification of novel leads and the development of new drug sources, because of their enormous scaffold diversity and structural complexity (Atanasov et al. 2021). Some plant-derived extracts and / or compounds, such as the alkaloids morphine and codeine from poppy (Papaver somniferum), and atropine isolated from Atropa belladonna, have been widely used in the treatment of well-known diseases (Calixto 2019). Also, in the past 30 years, many studies have focused on marine ecosystems, a reservoir of about 250,000–500,000 species of organisms, and revealed that the marine organisms also produce a wide range of structurally diverse compounds that act as a source of marine natural products for drug discovery (Khalifa et al. 2019, Lu et al. 2021). Especially, algae are rich sources in bioactive secondary metabolites and nutraceuticals such as carotenoids, polyphenols, and fatty acids with antioxidant, anti-inflammatory, antiviral, antibacterial, anti-obesity, atopic dermatitis regulation, and anticancer properties (Lee et al. 2021, 2022, Little et al. 2021, Menaa et al. 2021, Mihindukulasooriya et al. 2022, Pradhan and Ki 2023, Thakuri et al. 2023). Moreover, many recent studies regarding on aquaculture technique of various algae have been presented to develop the industrial utility technology of algal natural products (Choi et al. 2021, Kim et al. 2021, Hwang et al. 2022, Jiksing et al. 2022).
Fucoxanthin is a xanthophyll, which is a subset of carotenoids, produced by brown algae. Fucoxanthin exhibits anti-cancer (Martin 2015, Satomi 2017), anti-obesity (Gammone and D’Orazio 2015, Muradian et al. 2015), and anti-diabetic properties (Maeda 2015). In particular, fucoxanthin isolated from Sargassum siliquastrum—a brown alga abundant in Jeju Island, South Korea—showed protective effects against UV-B-induced cell damage (Heo and Jeon 2009) and H2O2-induced oxidative stress (Heo et al. 2008) and also exhibited anti-inflammatory effects (Heo et al. 2012). However, no studies have yet been conducted on the antiviral effect of fucoxanthin.
In this study, we investigated the antiviral effects of fucoxanthin isolated from S. siliquastrum to further explore the marine natural products available against coronavirus. The antiviral effects of fucoxanthin were confirmed against SARS-CoV-2-infected Vero cells. Structural analysis for the effects of fucoxanthin was fulfilled in a computational space and in vitro colorimetric method.
MATERIALS AND METHODS
Purification of fucoxanthin from Sargassum siliquastrum
Isolation and purification of fucoxanthin were carried out following a previous paper (Heo et al. 2012). In brief, S. siliquastrum, a brown alga, was collected along the Seongsan coast (33°27′32.9″ N, 126°56′31.2″ E) of Jeju Island, South Korea. Fucoxanthin was purified from S. siliquastrum through a series of steps, including 80% MeOH extraction, CHCl3 partitioning, silica column chromatography (silica gel, Merck, Darmstadt, Germany) Sephadex LH-20 column chromatography (Sephadex LH-20; Sigma Aldrich, Burlington, MA, USA), and reversed-phase high-performance liquid chromatography system (Alliance 2690; Waters Corporation, Milford, MT USA) (Supplementary Figs S1 & S2). The structure of fucoxanthin was identified by comparing the nuclear magnetic resonance spectrum data with those in the existing literature (Supplementary Figs S3 & S4). All solvents used were of analytical grade.
Virus and cells used
SARS-CoV-2 was provided by the Korea Centers for Disease Control and Prevention (KCDC; previously known as Korea Disease Control and Prevention Agency [βCoV/KOR/KCDC03/2020]). Vero cell line was purchased from the American Type Culture Collection (ATCC; ATCC-CCL81). Vero cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum and 1× antibiotic-antimycotic solution, and maintained at 37°C in a 5% CO2 incubator. SARS-CoV-2 was propagated in Vero cells. All in vitro experiments using SARS-CoV-2 were performed at the Institut Pasteur Korea, in compliance with the guidelines of the Korea National Institute of Health, using enhanced biosafety level 3 (BSL3) containment procedures in laboratories approved for use by the KCDC.
Analysis of antiviral activity of fucoxanthin
All in vitro experiments using SARS-CoV-2 were performed following a previous study (Jeon et al. 2020). Vero cells were seeded at 1.2 × 104 cells per well in black 384-well μ Clear plates (Greiner Bio-One, Chonburi, Thailand). After 24 h, fucoxanthin solution of concentrations ranging from 0.1 to 50 μM were added to the cells and incubated for 1 h. Then, SARS-CoV-2 was added to the wells at a multiplicity of infection of 0.0125, in the BSL3 containment facility. Antiviral activity was normalized to positive (mock) and negative (0.5% dimethyl sulfoxide) controls in each assay plate, and the values were measured in duplicate.
Preparation of 3D structure of fucoxanthin and the main proteins of SARS-CoV-2
For in silico studies, the 2D structure of fucoxanthin was obtained from PubChem (CID 5281239), and geometry optimization was performed using the prepare ligand and energy minimization protocols of Discovery Studio 2022 (Biovia, San Diego, CA, USA). The crystal structures of the main proteins of SARS-CoV-2 were obtained from a Protein Data Bank (PDB; http://www.pdb.org). ACE2-spike protein (PDB ID: 6LZG), main protease (Mpro) (PDB ID: 6LU7), and RNA-dependent RNA polymerase (RdRp; PDB ID: 7BV2), having a resolution of 2.50 Å, 2.16 Å, and 2.50 Å, respectively, were selected. Structure preparation of these proteins was conducted using the prepare protein protocol of Discovery Studio 2022 (Biovia). Each binding site of the main protein was defined, depending on the current docking site of its ligand, following a previous paper (Jin et al. 2020, Wang et al. 2020, Yin et al. 2020).
Molecular docking of fucoxanthin to the main proteins of SARS-CoV-2
Molecular docking analysis was performed to assess the respective binding positions and binding energies of fucoxanthin to the main proteins of SARS-CoV-2: ACE2-spike protein, Mpro, and RdRp. For this analysis, flexible docking based on the Chemistry at Harvard Macromolecular Mechanics (CHARMM) and ‘Calculate Binding Energies’ tools of Discovery Studio 2022 (Biovia) were used. The binding site of the proteins and fucoxanthin were allowed to move freely during the docking simulation. The docking positions of fucoxanthin on the main proteins were expressed as 3D crystal structures.
Molecular dynamic simulations of fucoxanthin complexes formed with the main proteins of SARS-CoV-2
To investigate the dynamic behavior of fucoxanthin–main protein complexes, molecular dynamic (MD) simulations were conducted using Discovery Studio 2022 (Biovia) based on the CHARMM force field. Each simulation step was designed based on the previous study (Jadhav et al. 2018, Jayaraman et al. 2021, Kang et al. 2023). The complex was solvated with explicit periodic boundary (orthorhombic box). Then, the solvated complex was first minimized for 1,000 steps using the steepest descent algorithm, and second minimized for 2,000 steps using adopted basis Newton-Raphson algorithm. And then, the minimized complex was heated for 100 ps at 300 K, equilibrated for 500 ps, and conducted production and nanoscale MD for 20 ns in standard number of particles, volume, and temperature ensemble and in a standard number of particles, pressure, and temperature ensemble. The stability of each complex was assessed by computing the root-mean-square deviation (RMSD) and fluctuation over the entire simulation period.
In vitro ACE2-spike protein binding inhibition effect
ACE2-spike protein inhibition effect of fucoxanthin was measured by using a SARS-CoV-2 S1 Protein-ACE2 Binding Inhibitor screening assay kit (Abcam PLC, Cambridge, UK) following the instructions in the enclosed user manuals. Briefly, 50 μL of fucoxanthin was added to the S1 protein-coated microplate for 30 min, and then 50 μL of the diluted biotinylated human ACE2 was mixed and incubated for 2 h. And then 100 μL of the diluted streptavidin-HRP was added to all of the wells and then incubated for 1 h. After incubation, 100 μL of TMB substrate was added to all wells and incubated for 20 min. Finally, 100 μL of stop solution was added to all wells, and the absorbance of each well was measured at 450 nm using Multiskan Go microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analysis
All in vitro data were represented as the mean ± standard deviation of three determinations. The statistical comparison was assessed by one-way ANOVA followed by Dunnett’s multiple comparison test using GraphPad Prism software version 9 (GraphPad Software, San Diego, CA, USA). Statistical significance was considered at p-values < 0.05 and 0.001.
RESULTS
Antiviral activity of fucoxanthin in SARS-CoV-2-infected Vero cells
Brown algae are known to be a good source of fucoxanthin; the antiviral activity of fucoxanthin derived from S. siliquastrum is shown in Fig. 1. Fucoxanthin inhibited the infection in a concentration-dependent manner, without showing any cytotoxicity. Fucoxanthin exhibited an inhibition rate of 61.45 ± 4.12% at 50 μM concentration.
Molecular docking of fucoxanthin to the main proteins of SARS-CoV-2
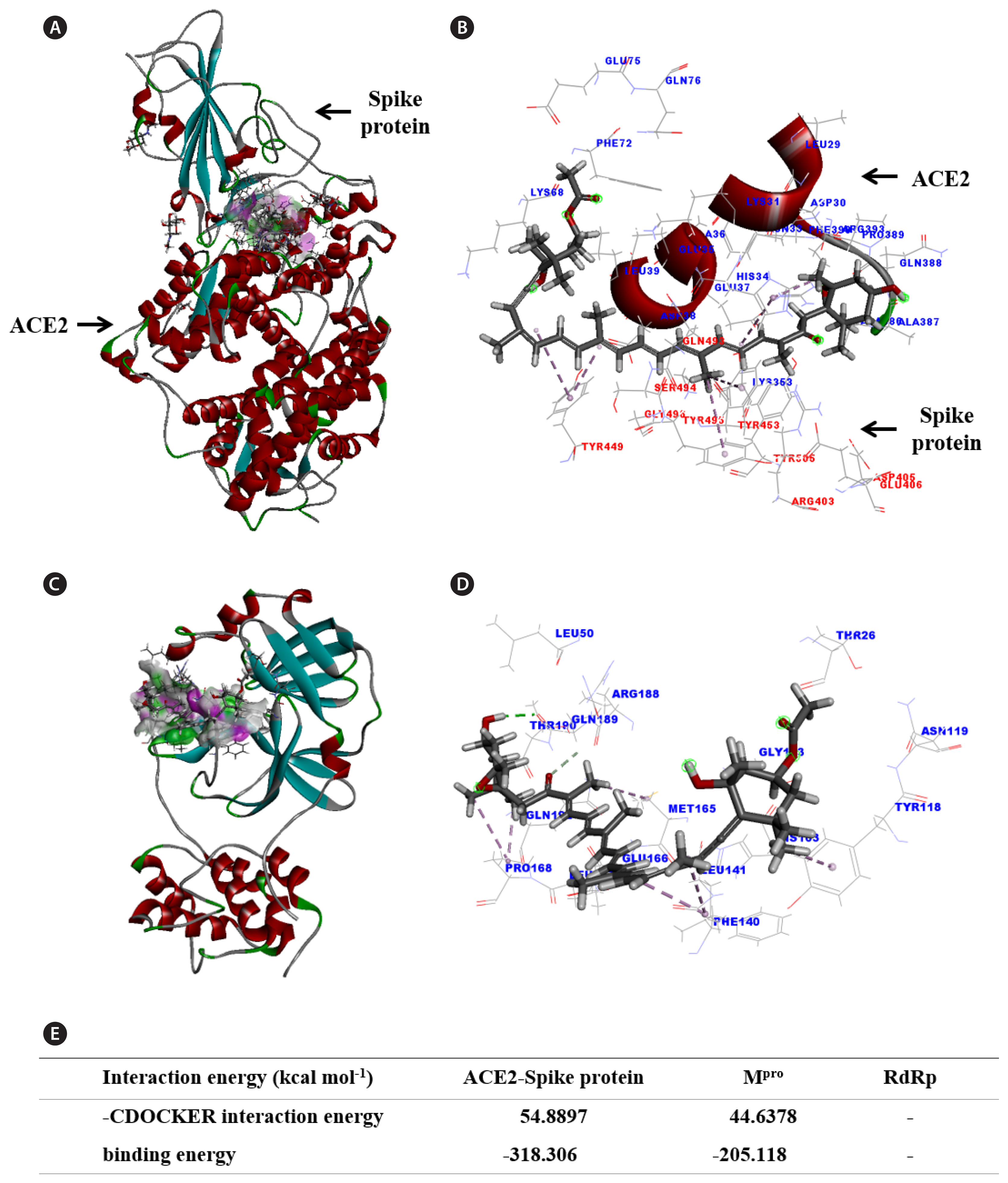
Molecular docking analysis of fucoxanthin was carried out by targeting the important main proteins in viral life cycle including ACE2-spike protein (viral entry), Mpro (processing polyproteins), and RdRp (replication of RNA) (Panda et al. 2020, Pandey et al. 2020, V’kovski et al. 2021, Kang et al. 2023). Results of molecular docking of fucoxanthin to the main proteins of SARS-CoV-2 are shown in Fig. 2. Fucoxanthin is displayed as a stick model and the main proteins of SARS-CoV-2, including ACE2-spike protein and Mpro, are displayed as solid ribbon models (Fig. 2A & C), as well as line models (Fig. 2B & D). Each binding site of fucoxanthin to the main proteins is shown in an acceptor–donor system (Fig. 2A & C).
Molecular docking of fucoxanthin to the ACE2-spike protein was performed by facilitating interactions with the binding site between ACE2 and the spike protein. The binding surface of the ACE2-spike protein was expressed by hydrogen bonds (Fig. 2A). The fucoxanthin-ACE2-spike protein complex displayed a favorable hydrogen bond with the donor (represented by the pink section) and the acceptor (represented by the green section) regions (Fig. 2A). The binding position of the fucoxanthin–ACE2-spike protein complex revealed a network of pi-alkyl bonds between the following residues: His34 (on ACE2), and TYR449, TYR453, and TYR495 (on spike protein) (Fig. 2B). The stability of the fucoxanthin-ACE2-spike protein complex was expressed as two types of energy values: the highest docking interaction energy (–CDOCKER interaction energy) was 54.8897 kcal mol−1, and the lowest total binding energy was −318.306 kcal mol−1 (Fig. 2E).
The fucoxanthin-Mpro complex also displayed a favorable hydrogen bond with the donor (represented by the pink section) and the acceptor (represented by the green section) regions (Fig. 2C). The binding position of the fucoxanthin-Mpro complex revealed a network of hydrogen and alkyl bonds between the following residues: Tyr118 (Pi-Alkyl), Leu141 (Alkyl), Met165 (Alkyl), Pro168 (Alkyl), Gln189 (conventional hydrogen bond and / or carbon hydrogen bond). The stability of the fucoxanthin-Mpro complex was expressed as two types of energy values: the highest docking interaction energy (–CDOCKER interaction energy) was 44.6378 kcal mol−1, and the lowest total binding energy was −205.118 kcal mol−1 (Fig. 2E). However, fucoxanthin failed to dock to RdRp (data not shown). These results indicated that fucoxanthin combined more strongly with the ACE2-spike protein, compared to Mpro, and has the least potential for docking to RdRp.
MD simulation of fucoxanthin on ACE2-spike protein and Mpro
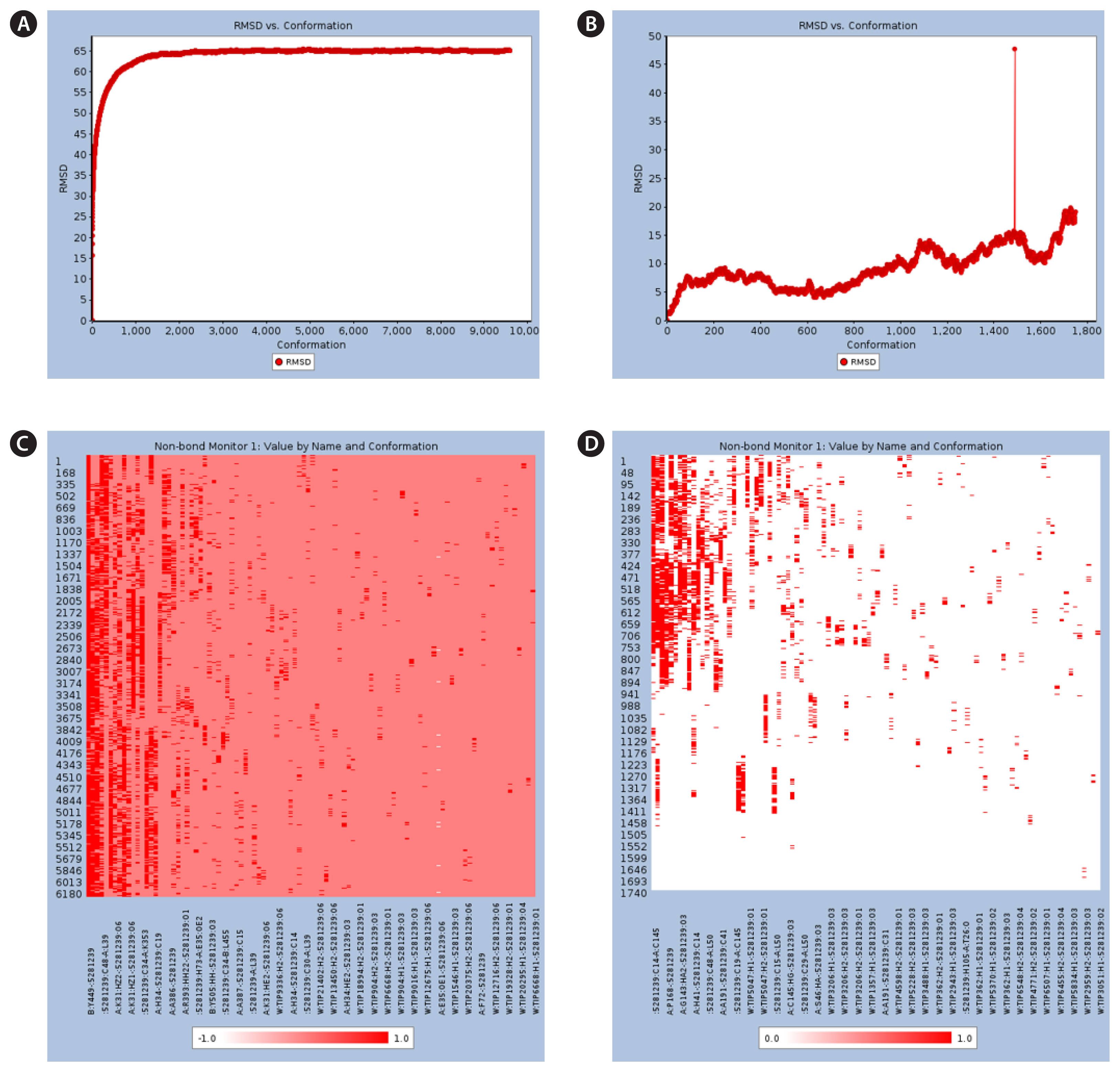
The stability of each of the fucoxanthin complexes was predicted using MD simulations. Fucoxanthin remained continuously docked to the active site of the ACE2-spike protein at the end of a 20 ns simulation (Fig. 3A), whereas the docking broke with Mpro in the middle of the simulation (Fig. 3B). Furthermore, the total energy of the conformations of fucoxanthin-ACE2-spike protein complex was found to be relatively unchanged for a duration of 1.5–10 ns (Fig. 3C), whereas the total energy of the conformations of fucoxanthin-Mpro complex was reduced, until the breakout of fucoxanthin (Fig. 3D).
Moreover, RMSD analysis of the conformations on the initial frame in the trajectory indicated that fucoxanthin achieved a stable conformation with ACE2-spike protein for up to a 20 ns simulation (Fig. 4A); whereas, fucoxanthin formed an unstable conformation with Mpro, showing the jumping value at about 3 ns (Fig. 4B). In the fucoxanthin-ACE2-spike protein complex, the Tyr449 residue on the spike protein was essential for stability of the complex, which was demonstrated by a sustainable interaction at a frequency of 1.149 (Fig. 4C, Supplementary Table S1). Although some amino acids on Mpro, such as Pro168, did form interactions with fucoxanthin, the interactions broke in the middle of the simulation (Fig. 4D, Supplementary Table S2). These MD simulation results indicated that fucoxanthin could form strong and stable complexes with the ACE2-spike protein, but not with Mpro.
In vitro analysis of ACE2-spike protein binding inhibition effect of fucoxanthin
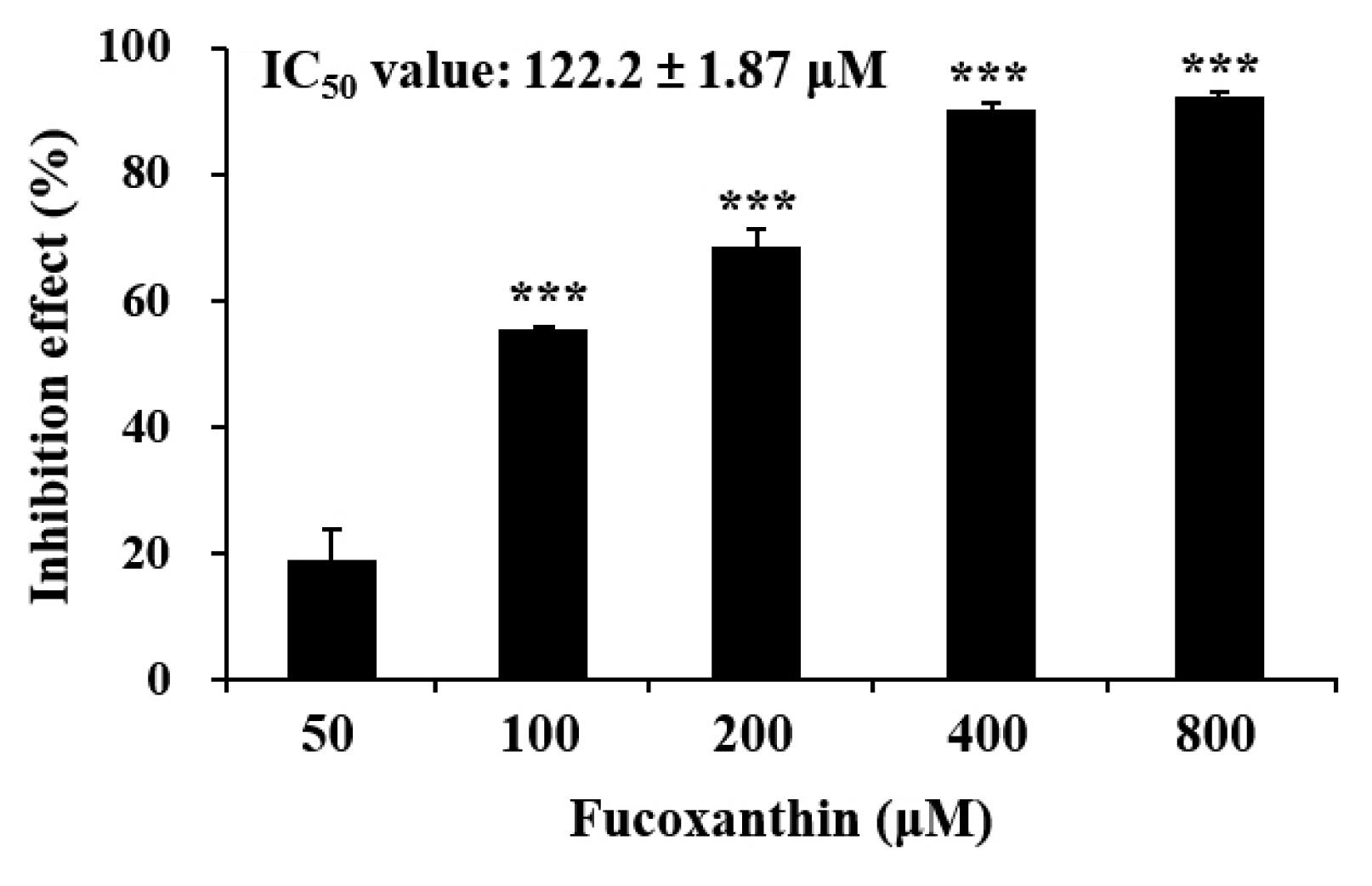
To confirm the molecular docking and simulation prediction results on the fucoxanthin–ACE2-spike protein complex, in vitro ACE2-spike protein binding inhibition effect of the fucoxanthin was evaluated. The fucoxanthin inhibited the binding between ACE2 and spike protein in a concentration-dependent manner with the range of 50–400 μM, and the IC50 value were calculated to be 122.2 ± 1.87 μM (Fig. 5). This result verified that the fucoxanthin possessed the antiviral effect by inhibiting ACE2-spike protein binding.
DISCUSSION
The imbalances related to oxidative stress and inflammation in the host cells may, in turn, be responsible for viral infections; therefore, inflammatory mediators are correlated to oxidative stress in the infected host cells (Zeremski et al. 2007, Pereira et al. 2021). Interestingly, the formation of reactive oxygen species (ROS) and hyper-inflammation, due to factors such as cytokine storm syndrome, are both major predictors of the development, severity, and mortality of coronavirus disease-19 (Katsiki and Ferrannini 2020, Tan et al. 2021, Vollbracht and Kraft 2022). Thus, many researchers have attempted to explore the antiviral properties of compounds with antioxidant and anti-inflammatory properties (Katsiki and Ferrannini 2020, Rehman et al. 2021, Tan et al. 2021, Pisoschi et al. 2022). According to the results of previous studies, fucoxanthin derived from S. siliquastrum decreased intracellular ROS generation and cell damage caused by exposure to UV-B radiation or hydrogen peroxide (Heo et al. 2008, Heo and Jeon 2009). Also, fucoxanthin inhibited the production of inflammatory mediators such as nitric oxide, prostaglandin E2, tumor necrosis factor-α, and interleukin-6 (Heo et al. 2012). Furthermore, in this study, fucoxanthin inhibited the infection of SARS-CoV-2 in Vero cells in vitro. Fucoxanthin shows both antioxidant and anti-inflammatory properties, as well as antiviral activities.
Molecular docking is a prediction algorithm for determining the binding affinities between a small molecule and a protein target (Torres et al. 2019, Kang et al. 2022). Molecular dynamics is a computer simulation method for evaluating the stability of a small molecule binding to a protein target (Guterres and Im 2020). Several previous studies have reported on ligand-protein complex formations by validating their results using molecular docking and MD simulations, to support their in vitro results (Elkaeed et al. 2022, Saleem et al. 2022, Deswal et al. 2023, Nassar et al. 2023).
The virus-specific molecular interaction with the host cell can be a key target for exploring antiviral drugs. SARS-CoV-2 enters the host cell by binding the spike protein to cellular ACE2 receptor (Choudhary et al. 2020, Panda et al. 2020). Mpro, a cysteine protease residing in nsp5, is responsible for the proteolysis of processing of major polyproteins cleavage sites resulting in the formation of viral nonstructural proteins. Mpro is considered a promising drug target because Mpro is dissimilar to human protease while it is closely related to those of other betacoronaviruses (Dai et al. 2020, Khan et al. 2020, Ullrich and Nitsche 2020).
In silico studies on the fucoxanthin-main protein complexes showed that fucoxanthin combined with the ACE2-spike protein by revealing a network of pi-alkyl bonds with some amino acid residues such as Tyr449, Tyr453, and Tyr495 (on the spike protein). Also, the results of the MD simulations indicated that fucoxanthin could form strong and stable complexes with the ACE2-spike protein, but not with Mpro. Particularly, Tyr449 is an essential amino acid for the stability of the fucoxanthin-ACE2-spike protein complex. Tyr449 and Tyr453 are major amino acids of the spike protein that bind to Umifenovir, a well-known inhibitor of ACE2-spike protein (Choudhary and Silakari 2020). This finding suggests that fucoxanthin inhibits SARS-CoV-2 by acting similarly to Umifenovir.
SARS-CoV-2 infection causes renin angiotensin system activation containing ACE2. ACE1 metabolizes angiotensin I to angiotensin II, thus leading to increased vasoconstriction. While ACE2 leads vasodilation by changing angiotensin II produced by ACE1 to angiotensin 1–7. It means that the binding of SARS-CoV-2 to ACE2 inhibits ACE2 function, and results in disturbance of vasodilation (González-Rayas et al. 2020). Previous study presented the inhibition effect of fucoxanthin from Sargassum sp. on ACE in in vitro and in silico (Raji et al. 2020). Also, in this study, fucoxanthin possess antiviral activity against SARS-CoV-2 by inhibiting the binding of spike protein to ACE2. These results suggested that fucoxanthin possess both blood pressure control effects and SARS-CoV-2 infection inhibition effects. Thus, further investigations are required to verify the multiple effect of fucoxanthin as a therapy for SARS-CoV-2 infection and / or the vascular disease by the infection.
According to a previous study, fucoxanthin displays low toxicity in normal cells both in vitro and in vivo (Peng et al. 2011). Furthermore, the U.S. Food and Drug Administration (FDA) has acknowledged the use of fucoxanthin as a dietary ingredient via a New Dietary Ingredient notification of fucoxanthin (FDA, updated 2017). Thus, fucoxanthin can be a new potential drug candidate. More detailed studies are required to establish the efficacy and optimal concentration of fucoxanthin use against SARS-CoV-2, as well as to determine its safety and bioavailability in in vivo models. Furthermore, in order to increase the industrial utility of algal natural products including fucoxanthin, research on the quantitative-structure activity relationships of algal natural products should be conducted through the accumulation of training set data based on artificial intelligence technology.
In conclusion, to determine the antiviral activity of fucoxanthin derived from S. siliquastrum, we conducted in vitro tests using SARS-CoV-2-infected Vero cells, in silico confirmations using molecular docking and MD simulations, and in vitro verification on the structural bond using colorimetric method. Fucoxanthin exhibited an inhibition rate of 61.45 ± 4.12% at 50 μM concentration. On the basis of this observation, along with the results of in silico docking of fucoxanthin to ACE2-spike protein and in vitro inhibiting on the binding between ACE2 and spike protein, it was verified that the antiviral activity of fucoxanthin might be due to the blocking of viral entry. Therefore, fucoxanthin can be a potential candidate for the treatment and / or prevention of coronaviruses, including SARS-CoV-2.