ABSTRACTThe marine brown algal genus Colpomenia has a worldwide distribution, with five species reported in Korea and Japan. However, no studies to date attempted to identify the number of species and geographical distribution of Colpomenia along Chinese coast. To fill the biodiversity knowledge gap, we analyzed 63 mitochondrial cox3 and 62 mitochondrial atp6 sequences of Colpomenia specimens collected from 30 localities along the Chinese coast. Maximum likelihood and Bayesian inference trees suggest the presence of at least three Colpomenia species (i.e., C. peregrina, C. claytoniae, and C. sinuosa) in China. C. peregrina and C. claytoniae are documented for the first time. C. sinuosa was only found in the South China Sea and its distribution didn’t overlap with that of C. peregrina which was found in the Yellow-Bohai Sea and the East China Sea. C. claytoniae appears to be confined to three isolated islands in the East and the South China Sea, where it occurs in sympatry with, respectively, C. peregrina and C. sinuosa. Future study can focus on comparing eco-physiological differences of Colpomenia species in response to environmental variables and exploring possible genetic hybridization/introgression at inter-specific contact zones.
INTRODUCTION
Colpomenia (Endlicher) Derbès et Solier (Scytosiphonaceae, Phaeophyceae) is a cosmopolitan brown algal genus frequently found attached to rock with gentle slopes or attached to other marine macroalgae at middle or lower intertidal zone. Due to developmental variation, Colpomenia has been traditionally divided into two morphological groups: (1) globular species producing plurilocular and unilocular zoidangia on crustose sporophytes, and (2) species with elongate or tubular thalli producing only unilocular zoidangia on sporophytes (Kogame et al. 1999, Boo et al. 2011, Lee et al. 2012). However, Santiañez et al. (2018) recently removed the tubular species [i.e., Colpomenia bullosa (D. A. Saunders) Yamada, Colpomenia durvillei (Bory) M. E. Ramírez, and Colpomenia wynnei K. M. Lee, R. Riosmena-Rodriguez, K. Kogame et S. M. Boo] from Colpomenia and proposed them as a new genus Dactylosiphon Santiañez, K. M. Lee, S. M. Boo et Kogame based on combined mitochondrial and chloroplast phylogenies.
The globular Colpomenia species usually contain oxygen generated by photosynthesis, making them buoyant. When oysters are the substrate, they can become buoyant and drift with the current, earning members of the globular species the moniker “oyster thieves” (Blackler 1967, Vandermeulen 1984). Adding to such mischief caused by the globular species, floating thalli can travel long distances and spread Colpomenia species in oyster mariculture (Lee et al. 2014a). Therefore, the behavior and mobility of globular Colpomenia species make them ideal for studying how maritime transportation affects biodiversity trends, population genetic structure and biogeographical patterns of seaweeds.
Ten Colpomenia species have been recognized (Guiry and Guiry 2019). Five species have been recorded from the coasts of Korea and Japan, i.e., C. claytoniae S. M. Boo, K. M. Lee, G. Y. Cho et W. Nelson (Boo et al. 2011), C. ecuticulata M. J. Parsons (Lee and Kang 2001), C. expansa (D. A. Saunders) Y. -P. Lee (Lee 2008, Lee et al. 2012), C. peregrina Sauvageau (Kogame et al. 1999, Boo 2010), and C. sinuosa (Mertens ex Roth) Derbès et Solier (Boo 2010, Yoshida et al. 2015). These studies indicate that Colpomenia species diversity in the northwest Pacific is rich. C. sinuosa is common in Korea, Japan, and China (e.g., Okamura 1936, Tseng 1983, Kogame 1997, Cho et al. 2005, Lee et al. 2013). Cho et al. (2009) presented a first glimpse at genetic diversity of C. sinuosa on a global scale. They analyzed nuclear internal transcribed spacers of the ribosomal cistron (ITS) and chloroplast encoded large subunit of the ribulose-1,5-bisphosphate carboxylase gene (rbcL) and found that the unique tandem repeats in the ITS region correspond to the geographical distribution of C. sinuosa. Lee et al. (2013) observed three genetic lineages within C. sinuosa around the world using mitochondrial cytochrome c oxidase subunit III (cox3) and chloroplast rbcL. They suggested that C. sinuosa had broad and complicated biogeographical patterns in the Indo-Pacific Ocean region where the Red Sea was likely a refugium before the Last Glacial Maximum (LGM), and multiple introductions might occur among Australasia, Europe, and America.
Chinese coast, in particular the Yellow-Bohai Sea (YBS), geographically adjoins and shares homologous coastal marine environments with the Korean Peninsula and the Japanese Archipelago (Tyberghein et al. 2012). Similar coastal configurations in those neighboring areas during the LGM (Wang 1999), produced seaweed floras that are closely related taxonomically (Tseng 1983, Yoshida et al. 2015). In addition, the Kuroshio Current can connect populations of marine species, including seaweeds, along the coasts of East Asia (Barkley 1970, Hu et al. 2013, Li et al. 2017). The coastal currents in China (e.g., the Bohai Sea coastal current, the Yellow Sea coastal current and the East China Sea coastal current) in different seasons can also contribute to multiple dispersal to a great extent. Taken together, these historical climate change and oceanic currents can interplay and ultimately affect the richness, diversity and distribution patterns of present-day seaweeds in China, Korea, and Japan (e.g., Chondrus ocellatus Holmes, Hu et al. 2015, Sargassum fusiforme (Harvey) Setchel, Hu et al. 2017).
Colpomenia species have broad latitudinal distributions in the West Pacific (Guiry and Guiry 2019). For example, C. peregrina appears in coastal temperate waters in the northern hemisphere of Japan, Korea, and Russia (e.g., Perestenko 1980, Lee et al. 2014a, Yoshida et al. 2015) and in the southern hemisphere of southern Australia and New Zealand (e.g., Harper et al. 2012, Nelson 2013, Scott 2017), respectively. Similar patterns have been reported for C. claytoniae (e.g., Boo and Ko 2012, Nelson 2013). China, which stretches over some 18,000 kilometers of coastlines and has considerable environmental shifts (e.g., the significantly different sea surface temperature, annual precipitation and solar radiation between northern and southern coast) (Ma et al. 2017), is the main component of the China Sea Coastal Province (Longhurst 2007) and geographically located near Japan and Korea. However, for the genus Colpomenia, only C. sinuosa has been reported in the past few decades (Tseng 1983, 2008, Liu 2008), and there has been no study of the quantity and distribution of Colpomenia species. We thus speculate that previous studies (Tseng 1983) may have underestimated the number of Colpomenia species along the coast of mainland China. Furthermore, climate change induced a northward shift of temperature zones in China, together with ocean acidification and coastal erosion, have exerted the necessary obligation of protecting seaweed species diversity (Zheng et al. 2018). The present study focuses on Colpomenia specimens collected from the entire Chinese coast. We conducted morphological observations and phylogenetic reconstructions using mitochondrial cox3 and atp6 genes to determine species diversity and phylogenetic relationships in Colpomenia and range limits along the coast of China. This information may provide useful insights into discovering and conserving natural seaweeds resources in a time of climate change.
MATERIALS AND METHODSSampling and DNA extraction
Colpomenia specimens were collected from 30 coastal localities, from 18.21° N, 109.51° E to 39.02° N, 122.73° E (Fig. 1, Supplementary Table S1). At each site, 1–3 specimens were collected, with an interval transect ≥10 m to avoid sampling genetically homogeneous plants. Specimens were cleaned from epiphytes, washed and partially preserved in silica gel. Twenty specimens chosen randomly for morphological sections were kept in 4% formaldehyde and deposited in the Marine Biological Herbarium of the Institute of Oceanology, Chinese Academy of Sciences (voucher numbers 2017BC0015-2017BC0034).
Total genomic DNA was extracted using the DNAsecure Plant Kit (TIANGEN, Beijing, China) following the manufacturer’s instructions. Purity and quality of genomic DNA were checked by OD260/OD280 ratios and 1% agarose gel electrophoresis.
Polymerase chain reaction amplification and DNA sequencingBased on a recent molecular study of Colpomenia (Lee et al. 2014a), mitochondrial cox3 and atp6 were selected as gene markers. Primer sets F49/R20 and F25P/R754P (Lee et al. 2014a) were used to amplify, respectively, cox3 and atp6 fragments from 63 specimens. Polymerase chain reaction (PCR) amplification was performed in 50 μL volumes, each containing 25 μL 2× Es Taq Master Mix (+dye) (CWBIO, Beijing, China), 10 pmol of each primer and 6 ng of template DNA. For the cox3 gene, PCR reactions consisted of an initial denaturation at 95°C for 4 min, followed by 35 amplification cycles (denaturation at 94°C for 45 s, annealing at 53°C for 1 min, extension at 72°C for 1 min) with a final extension at 72°C for 10 min. For the atp6 gene, PCR reactions consisted of an initial denaturation at 95°C for 4 min, followed by 35 amplification cycles (denaturation at 94°C for 30 s, annealing at 45°C for 30 s, extension at 72°C for 1 min) with a final extension at 72°C for 10 min. Quantity and quality of PCR products were checked using 1% agarose gel electrophoresis. Purification and sequencing of PCR products were performed using a BigDye Terminator Cycle sequencing kit and an ABI3730 automated sequencer (Applied Biosystems, Foster City, CA, USA).
Phylogenetic analysisTwenty-five recently published cox3 sequences and 6 atp6 sequences of Colpomenia (Boo et al. 2011, Lee et al. 2012, 2013, 2014a, 2014b) (Supplementary Table S2) were retrieved from GenBank to determine gene boundaries. Newly generated cox3 and atp6 fragments were aligned with referenced sequences using the MUSCLE model in MEGA v7.0 (Kumar et al. 2016). After editing and removal of excess bases, aligned sequences were adjusted manually to avoid false gaps.
Phylogenetic relationships were reconstructed based on the cox3 alignment, the atp6 alignment, and a concatenated cox3 + atp6 alignment, respectively using both maximum likelihood (ML) and Bayesian inference (BI). JModelTest v2.1.7 (Darriba et al. 2012) was used to determine the best-fit substitution model through Bayesian Information Criterion for each gene marker separately (cox3: TPM1uf + I + G [I = 0.35, G = 0.47], atp6: HKY + G [G = 0.14]). ML trees based on cox3 and atp6 were reconstructed in PhyML v3.1 (Guindon and Gascuel 2003) using 1,000 bootstrap replicates with a Nearest-Neighbour-Interchange heuristic method. The ML tree based on concatenated cox3 + atp6 alignment was reconstructed in RAxML v8.0 (Stamatakis 2014). BI trees were reconstructed in MrBayes 3.2 (Ronquist et al. 2012), running four Markov chain Monte Carlo iteration chains simultaneously for 107 generations, keeping one tree every 100 generations, with the first 104 trees sampled discarded as ‘burn-in.’ Consensus topology and posterior probability (PP) values were calculated for the remaining trees. The cox3 and atp6 fragments of Ectocarpus siliculosus (Dillwyn) Lyngbye (Cock et al. 2010) and Scytosiphon lomentaria (Lyngbye) Linkwere (Liu et al. 2016) were used as outgroup. Trees were visualized using FigTree v1.4.3, available at http://tree.bio.ed.ac.uk/software/figtree/.
Genetic variationDnaSP v5.10 (Librado and Rozas 2009) was used to count variable sites and parsimony informative sites in each marker. Using 1,000 bootstrap replications, genetic distances among 30 sampling sites were computed using MEGA v7.0, with a p-distance substitution model including transitions and transversions. A heatmap and a boxplot were used to present intra- and inter-specific genetic distances, respectively.
Morphological observationMultiple thalli were each cut at multiple parts and then were cut into thin slices of about 2–5 μm using a razor blade by hand and freezing microtome HM505E (MICROM, Neuss, Germany). Sections were stained in 1% aniline blue for 5–10 min, followed by decolorization with one or two drops of 1 M hydrochloric acid for 30 s and washed with seawater (Aisha and Shameel 2012). Sections were mounted and sealed in a mixture of 75% glycerin with aniline blue, observed and photographed under an optical microscope BX51TF (Olympus, Tokyo, Japan) and a fluorescence microscope Eclipse Ni (Nikon, Tokyo, Japan).
RESULTSMolecular diversity of mitochondrial cox3 and atp6Sixty-three cox3 sequences with an aligned length of 620 bp yielded 102 variable sites and 94 parsimoniously informative sites. Sixty-two atp6 sequences with an aligned length of 611 bp contained 107 variable sites and 103 parsimoniously informative positions.
Species diversity of Colpomenia along the coast of ChinaThe Colpomenia phylogenetic relationship based on the cox3, atp6 and concatenated cox3 + atp6 datasets were congruent (Fig. 2, Supplementary Figs S1–S4). The specimens from China were represented in three distinct species clades. The ML and BI trees both indicated that five specimens from Weizhou Island, Guangxi (WZ), Huokun’ao, Zhejiang (HK), and Dongfang, Hainan (DF) grouped with referenced C. claytoniae, forming a unique clade with well-supported values [cox3 + atp6: ML bootstrap percentage (BP) = 100, BI PP = 1.00; cox3: BP = 95, PP = 1.00; atp6: BP = 99, PP = 1.00]. Thirty-two sequences from Weizhou Island, Guangxi (BG, ST, HS, CT, and WZ), Guangdong (XW) and Hainan Island (WC, TO, CH, BA, DF, WN, LD, HW, HT, and DH) grouped with referenced C. sinuosa, forming another strongly supported clade (cox3 + atp6: BP = 100, PP = 1.00; cox3: BP = 99, PP = 1.00; atp6: BP = 100, PP = 1.00). The remaining 26 sequences from Liaoning (ZZ and HN), Shandong (CD, PL, GL, and SY), and Zhejiang (DL, LC, HJ, XH, HK, DS, and SM) grouped with the referenced C. peregrina and constituted the third clade (cox3 + atp6: BP = 76, PP = 1.00; cox3: BP = 94, PP = 1.00; atp6: BP = 98, PP = 0.94). It is therefore concluded that the three genetic clades found along the coast of China may correspond to C. claytoniae, C. sinuosa, and C. peregrina.
Genetic variations of cox3 and atp6 along Chinese coastPairwise genetic distances among sampling sites along Chinese coast based on cox3 and atp6 showed low genetic differentiations at the intra-specific level in Colpomenia (Fig. 3, Supplementary Tables S3–S5). Mitochondrial cox3 revealed a mean genetic distance of 0.5% within C. peregrina, 1.1% within C. sinuosa, and 1.9% within C. claytoniae (Supplementary Tables S3 & S5). Mitochondrial atp6 showed a mean genetic distance of 0.4% within C. peregrina, 0.5% within C. sinuosa, and 2.7% within C. claytoniae (Supplementary Tables S4 & S5). Much higher genetic differentiations were detected at the inter-specific level (Fig. 3, Supplementary Tables S3–S5). The cox3 gene showed a mean genetic distance of 10.3% between C. peregrina and C. sinuosa, 6.0% between C. peregrina and C. claytoniae, and 8.9% between C. sinuosa and C. claytoniae (Supplementary Table S3). Atp6 showed a mean genetic distance of 11.4% between C. peregrina and C. sinuosa, 6.0% between C. peregrina and C. claytoniae, and 12.8% between C. sinuosa and C. claytoniae (Supplementary Table S4).
Morphological characters of Colpomenia species
Table 1 indicates that morphological characters of the three Colpomenia species reported here (Fig. 4).
Colpomenia sinuosa (Mertens ex Roth) Derbès et Solier (Fig. 4A–D)Thalli attached erectly to substratum on rocks at intertidal and subtidal zone, irregularly hollow, saccate, 3–15 cm in diameter and yellowish brown in color (Fig. 4A). Phaeophycean hairs unicellular, about 100 μm long (Fig. 4B). Cortex consists of 4–6 layers of small polygonal cells and phaeoplasts (Fig. 4C). Medulla has five or more layers of irregular and large colorless cells (Fig. 4C). Sori discrete and punctate on thallus surface, with cuticle stained blue on plurilocular sporangia (Fig. 4C). Plurilocular sporangia cylindrical, uniseriate and biseriate, with cuticle stained blue, about 50 μm long (Fig. 4D); paraphysis brown in color, about 30 μm long (Fig. 4D). Unilocular sporangia were not observed.
Colpomenia claytoniae S. M. Boo, K. M. Lee, G. Y. Cho et W. Nelson (Fig. 4E & F)Thalli attached erectly on rocks at subtidal zone, globular or vesicle-like hollow sacs, irregularly convoluted and expanded or collapsed, 3–30 cm in diameter and yellowish green in color (Fig. 4E). Cortices have 1–2 layers of small polygonal cells with phaeoplasts (Fig. 4F). Medulla has 5–6 layers of increasingly larger, irregularly shaped cells (Fig. 4F). Sporangia and paraphyses were not observed.
Colpomenia peregrina Sauvageau (Fig. 4G–L)Thalli epilithic, thin, globular, 2–10 cm in diameter and greenish brown in color (Fig. 4G). Phaeophycean hairs start from medulla, 50–100 μm long (Fig. 4H). Cortical cells polygonal and irregularly arranged, 2–5 layers (Fig. 4I & J). Medullary cells large, irregular, and colorless, 2–4 layers (Fig. 4I & J). Plurilocular sporangia biseriate, club-shaped or elongate, about 30 μm long (Fig. 4K). Paraphysis club-shaped, as tall as plurilocular sporangia (Fig. 4L). Unilocular sporangia were not observed.
DISCUSSIONGenetic variations of cox3 and atp6 in ColpomeniaMitochondrial cox3 and atp6 have been previously used to identify new species in Colpomenia and examine genetic variation at inter- and intra-specific levels. In previous studies of the genus Colpomenia, cox3 indicated that C. peregrina differentiated from C. claytoniae by 4.40–7.69% (Boo et al. 2011, Lee et al. 2014a) and from C. expansa by 7.22% (Boo et al. 2011). Subsequently, cox3 was reported to differentiated by 12.9% between C. claytoniae and C. sinuosa (Lee et al. 2013). At the intra-specific level, cox3 shows 0–4.55% sequence divergence within C. claytoniae (Boo et al. 2011), 0–7.4% within C. sinuosa (Lee et al. 2013), and 0–3.7% within C. peregrina (Lee et al. 2014a). As for the atp6, the sequences up to 2.4% pairwise divergence within C. peregrina, and 4.8% between C. peregrina and C. claytoniae (Lee et al. 2014a). Furthermore, concatenation of cox3 and atp6 fragments shows average inter-specific divergence of 11.4% in Colpomenia and 0–2.9% in C. peregrina (Lee et al. 2014a).
In this study, mitochondrial cox3 and atp6 not only revealed diverse genetic distances between inter- and intra-clades in Colpomenia along the coast of China (Fig. 3, Supplementary Tables S3–S5), but also presented divergence values similar to the previous studies described above. This combined evidence further indicates that cox3 and atp6 are valuable markers for identifying hidden diversity in Colpomenia. Noticeably, the genetic distances of C. peregrina and C. sinuosa in cox3 were higher than those in atp6, while that of C. claytoniae in cox3 was lower than atp6. This kind of difference may result from the unequal evolutionary rate of cox3 and atp6. Additionally, the insufficiency of C. claytoniae sampling quantity could affect the accuracy of the results to some extent.
Colpomenia species along the coast of ChinaWidely distributed in temperate coastal waters of Korea and Japan (Lee and Kang 2001, Lee 2008, Boo 2010, Yoshida et al. 2015), C. peregrina Sauvageau was initially recognized as C. sinuosa var. peregrina due to its thin form, smooth bladders that are not folded or wrinkled and phaeophycean hairs more deeply seated than in C. sinuosa (Sauvageau 1927). Later, Hamel (1937) lifted it to species level. C. peregrina has extensive irregular sori without cuticles and thin thalli with 3–4 layers of colorless medullary cells (Clayton 1975, Cho et al. 2005). Nevertheless, distinguishing C. peregrina from C. sinuosa is challenging due to striking morphological similarity. In this study, morphological evidence allows us to distinguish three Colpomenia species mainly through the cuticle on the surface of plurilocular sporangia, the number of layers of outer cortex cells and medullary cells (Table 1).
C. claytoniae was newly identified based on congruent genetic evidence of rbcL and ITS regions and its distribution was considered to be restricted to Korea and Japan, and recently in Australia and New Zealand (Boo et al. 2011, Nelson 2013, Yoshida et al. 2015). It was characterized by a more irregular thallus with a deeply infolded surface. This species often appears epilithically in the lower intertidal, subtidal zones and tide pools (Boo et al. 2011).
Our combined morphological and phylogenetic evidence indicates that three Colpomenia species (i.e., C. peregrina, C. claytoniae, and C. sinuosa) occur in China. In addition, it is worthy to note that C. peregrina and C. claytoniae are recorded for the first time in China. Geographically, C. peregrina is distributed in the warm-temperate waters of the YBS and subtropical waters of the East China Sea; C. sinuosa is only distributed in the tropical coastal South China Sea, but the specific boundary between these two species has not been clear up to now. C. claytoniae may only appear in isolated islands in southern China. C. sinuosa and C. claytoniae overlap in distribution in WZ (Dishui village, Weizhou Island) and DF (Dongfang, Hainan Island); C. peregrina and C. claytoniae co-occur in HK (Huokun’ao, Nanji islands), but C. claytoniae grows at subtidal whereas C. peregrina grow at intertidal zone. Therefore, the geographical–scale distribution boundaries among Colpomenia species along the coast of China have not been delineated yet. Interestingly, only limited samples of C. claytoniae are reported in this study. But recent studies indicate that C. claytoniae occurs in the northern and southern West Pacific (e.g., Australia and New Zealand) (Boo et al. 2011, Nelson 2013), suggesting that extensive sampling is needed to identify the complete distribution range of each Colpomenia species along the Chinese coast. Furthermore, concatenated cox3 + atp6 revealed three genetic lineages in C. sinuosa (Fig. 2). Considering that Lee et al. (2013) reported multiple hidden lineages in this species, C. sinuosa may have cryptic diversity patterns and complicate trans-oceanic dispersal routes and evolutionary history.
Biogeographic processes contributing to Colpomenia diversityThe close relationships and co-distribution patterns of seaweed floras among the Yellow Sea, the South China Sea, the East Sea (Japan Sea) and the Pacific coast of Japan have been studied by Tseng and Chang (1963) over five decades ago. Such patterns can be ascribed to two main abiotic factors. First, LGM climate change caused sea levels to drop approximately 120 m and marginal seas in East Asia (e.g., the YBS and the East China Sea) disappeared completely (Guo 1979, Liu 1995, Wang 1999), which considerably impacted local natural coastal landscapes and resource allocation (Xie et al. 1996, Li and Chang 2009). Nevertheless, sea surface temperature, a vital factor shaping diversity and distribution patterns of seaweed (Müller et al. 2009, Tittensor et al. 2010), maintained the similar level in these marginal seas (Wang 1999). Consequently, seaweed floras in the YBS, the ECS and the Pacific coast of Japan underwent similar contraction-expansion processes induced by glacial and inter-glacial cycles, leading to many seaweed species co-distributed in China, Korea and Japan (Tseng and Chang 1959, van den Hoek 1984, Keith et al. 2014).
Second, oceanic current systems are of important factors in driving spatial dispersal and migration of seaweed (Tseng and Chang 1959). As illustrated in Fig. 1, the Kuroshio Current, which is the main system connecting the northern coast of China to other areas in the northwest Pacific, has been known to have a significant impact on species diversity and population connectivity (Barkley 1970, Hu et al. 2013, Lin et al. 2018). Therefore, the water flow driven by the Kuroshio Current can possibly promote trans-regional dispersal of Colpomenia species, expanding the geographical range from China to Korea and Japan.
Anthropogenic influence can also lead to seaweed introduction. Fouling of ships’ hulls, ballast water carried by trading vessels, and oyster mariculture are considered to be important pathways for the introduction of algae (Schaffelke et al. 2006). For example, Lee et al. (2014a) reported that C. peregrina was likely to be introduced from the NE Atlantic to the NW Atlantic via oyster mariculture and vessel hulls. Thus, maritime trade may contribute to the distributing expansion of Colpomenia species.
In summary, this study shows new occurrences of two Colpomenia species (C. claytoniae and C. peregrina) in China, with a clearly separate distribution between C. peregrina and C. sinuosa. The distribution range of subtidal C. claytoniae partially overlaps with intertidal C. peregrina and C. sinuosa in the East and South China Sea (Fig. 1). These contrasting distribution patterns inspire further exploration of how eco-physiological differences of Colpomenia species respond to environmental shifts, population-level phylogeographical diversification and evolutionary history, including possible genetic hybridization/introgression at inter-specific contact zones.
ACKNOWLEDGEMENTSWe would like to thank Drs. Li Jing-jing and Zhang Jie and PhD Ruo-Yu Liu (IOCAS) for the help in field collections. Special thanks go to Donald Sturge for carefully checking and improving the English writing. The study was supported by a joint research grant between National Natural Science Foundation of China (41761144057) and Thailand Research Fund (RDG6130002).
SUPPLEMENTARY MATERIALSSupplementary Table S1.Sampling details of Colpomenia in this study (https://www.e-algae.org). Supplementary Table S2.Mitochondrial cox3 and atp6 sequences retrieved from GenBank in this study (https://www.e-algae.org). Supplementary Table S3.MtDNA cox3 based mean genetic divergences among three Colpomenia species along the coast of China (https://www.e-algae.org). Supplementary Table S4.MtDNA atp6 based mean genetic divergences among three Colpomenia species along the coast of China (https://www.e-algae.org). Supplementary Table S5.MtDNA cox3- and atp6-based genetic divergences among sampling sites of Colpomenia along the coast of China (https://www.e-algae.org). Supplementary Fig. S1.The maximum likelihood (ML) tree derived from cox3 sequences of Colpomenia. Numbers above or near branches are ML bootstrap values. Values <50% were not shown (https://www.e-algae.org). Supplementary Fig. S2.The Bayesian inference (BI) tree derived from cox3 sequences of Colpomenia. Numbers above or near branches are BI posterior probabilities. Values <0.50 were not shown (https://www.e-algae.org). Supplementary Fig. S3.The maximum likelihood (ML) tree derived from atp6 sequences of Colpomenia. Numbers above or near branches are ML bootstrap values. Values <50% were not shown (https://www.e-algae.org). Supplementary Fig. S4.The Bayesian inference (BI) tree derived from atp6 sequences of Colpomenia. Numbers above or near branches are BI posterior probabilities. Values <0.50 were not shown (https://www.e-algae.org). Fig. 2Phylogenetic trees derived from concatenated cox3 + atp6 sequences of Colpomenia. Numbers above or near branches are maximum likelihood bootstrap values (left)/Bayesian inference posterior probabilities (right). Values <50% or <0.50 were not shown. 
Fig. 3Heatmap (A) and boxplot (B) showing average genetic distances using cox3 and atp6 genes among sampling sites, calculated by p-distance model (1,000 bootstrap replications). Each value is visualized as a square color-block in the heatmap, varying from 0 to 0.125. Lower-left blocks represent values of cox3 and upper-right blocks represent those of atp6. The red boxes in the boxplot represent genetice distances using cox3 gene while the black boxes represent genetic distances using atp6 gene. C., Colpomenia.
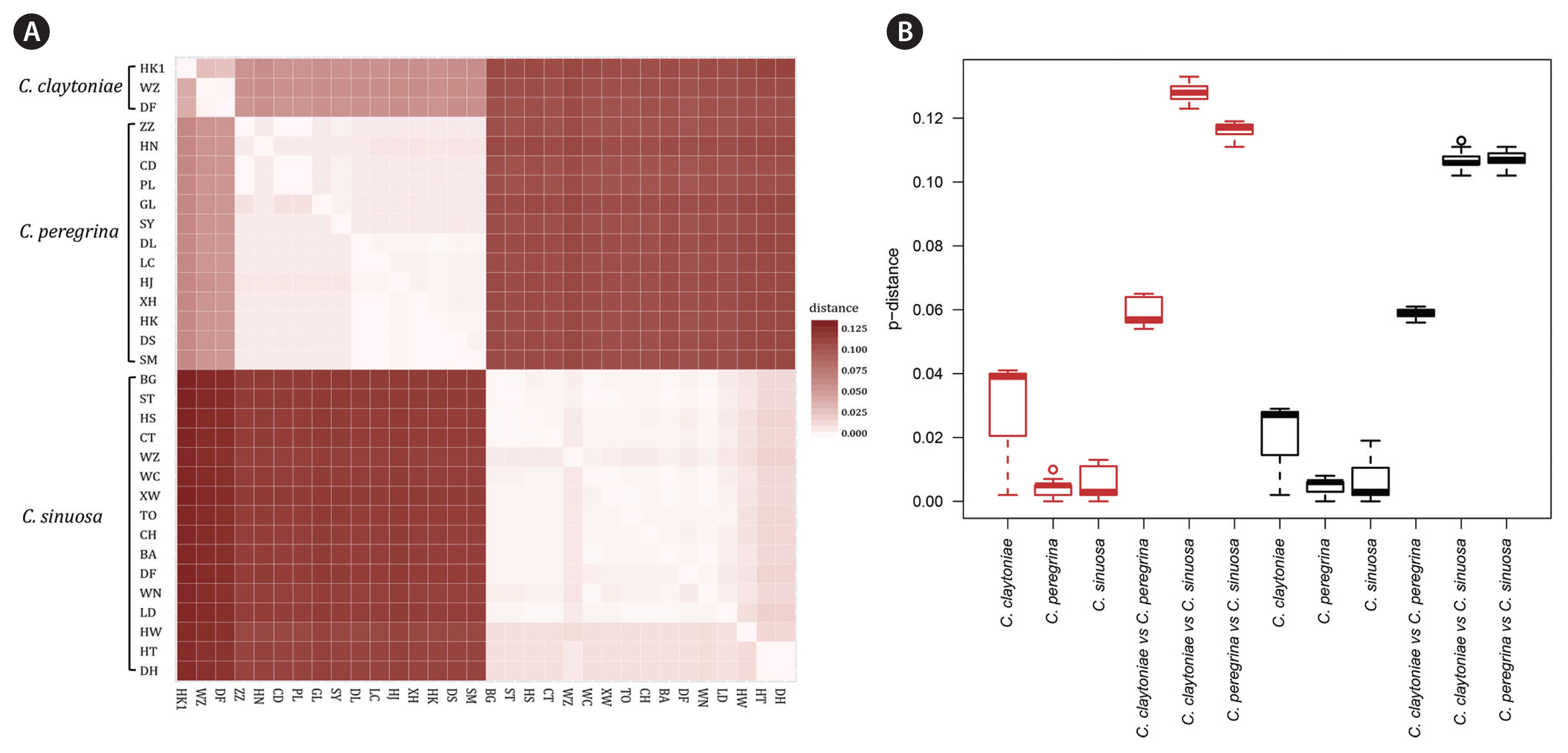
Fig. 4Morphology of Colpomenia species. (A) Colpomenia sinuosa thallus growing attached to rocks above the lowest tide (collected from BA in Hainan, Apr 14, 2018). (B) Phaeophycean hairs (black arrow) arising from cortical cells (black arrowhead), and medulla (white arrowhead) of C. sinuosa (sample collected from CH in Hainan, Mar 31, 2019). (C) Sori growing on the surface (black arrows) with cuticle (stained blue), cortex (black arrowhead), and medulla (white arrowhead) of C. sinuosa. (D) Close-up view of plurilocular sporangia (arrow) and paraphysis (arrowhead). (E) Colpomenia claytoniae thallus growing attached to rocks in subtidal zone, about 5 m below the lowest tide (collected from HK in Nanji islands, May 15, 2018). (F) Cortex (arrow) and medulla (arrowhead) of C. claytoniae. (G) Colpomenia peregrina thallus growing attached to rocks about 1.5 m above the lowest tide (collected from DS in Nanji islands, May 16, 2018). (H) Paraphysis (arrow) arising from medullary cells, and nascent plurilocular sporangia (arrowhead) of C. peregrina (sample collected from SY in Shandong, May 20, 2019). (I) Cortex (arrowhead) and medulla (arrow) of C. peregrina. (J) Close-up view of cortex (arrow) and medulla (arrowhead) of C. peregrina. (K) Plurilocular sporangia stained blue (arrow) of C. peregrina. (L) Paraphysis in blue (arrows) of C. peregrina. Scale bars represent: A, E & G, 1 cm; B, D, F & J, 50 μm; C & I, 100 μm; H, K & L, 10 μm. [Colour figure can be viewed at http://www.e-algae.org]. 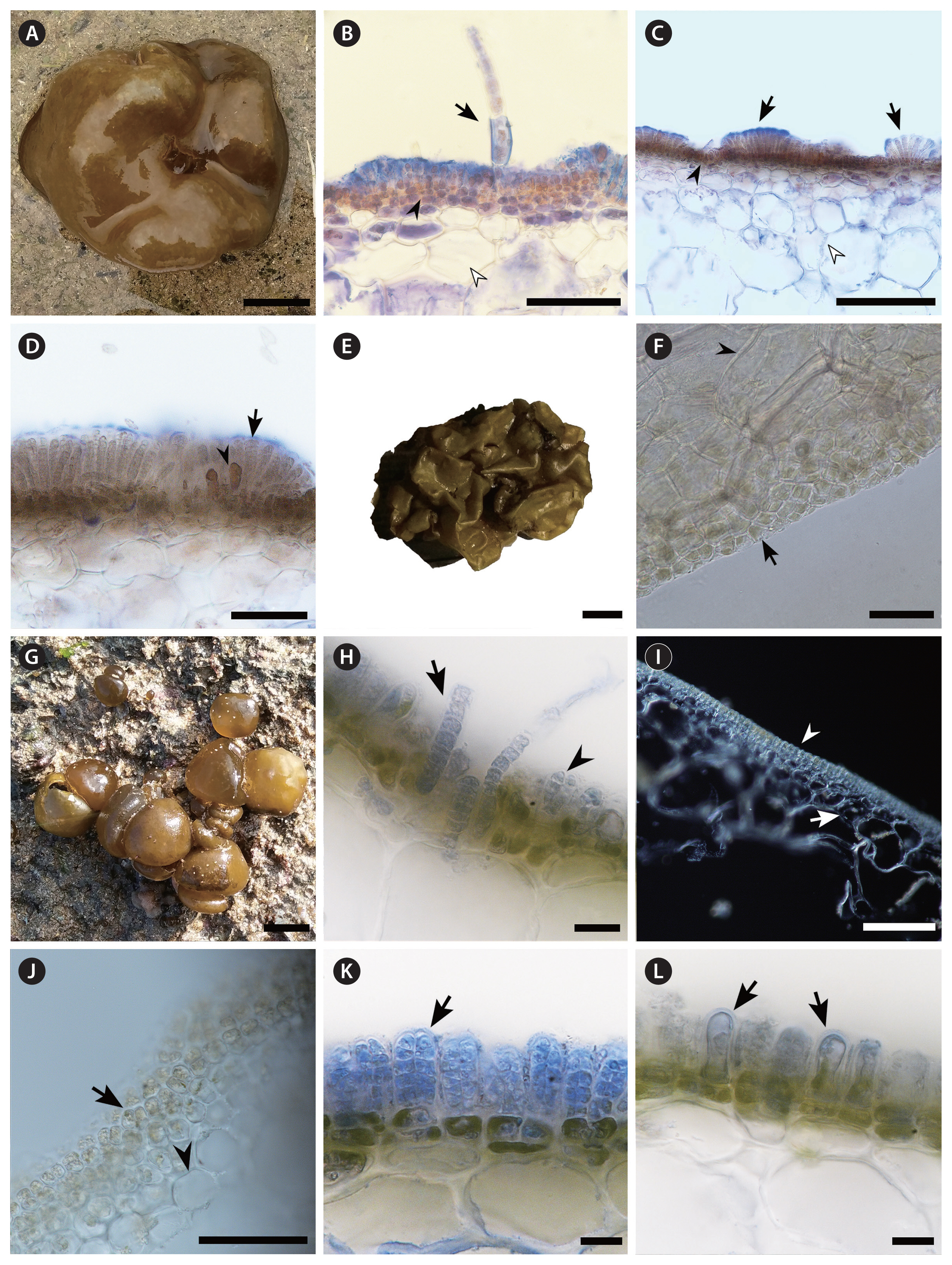
Table 1Morphological characteristics of three Colpomenia species along the coast of China
REFERENCESAisha, K. & Shameel, M. 2012. Taxonomy of the genus Colpomenia (Laminarophyceae, Phaeophycota) from the coast of Karachi. Proc Pak Acad Sci. 49:123–129.
Barkley, RA. 1970. The Kuroshio current. Sci J. 6:54–60.
Blackler, H. 1967. The ocurrence of Colpomenia peregrina (Sauv.) Hamel in the Mediterranean (Phaeophyta: Scytosiphonales). Blumea. 15:5–8.
Boo, SM. 2010. Scytosiphonaceae, Petrospongiaceae. In : Kim HS, Boo SM, editors Algal Flora of Korea. Vol. 2, No. 1. Heterokontophyta: Phaeophyceae: Ectocarpales. Marine Brown Algae 1. National Institute of Biological Resources, Incheon, 155–185.
Boo, SM. & Ko, YD. 2012. Marine plants from Korea. Marine & Extreme Genome Research Centre Program, Seoul, 233 pp.
Boo, SM., Lee, KM., Cho, GY. & Nelson, W. 2011.
Colpomenia claytonii sp. nov. (Scytosiphonaceae, Phaeophyceae) based on morphology and mitochondrial cox 3 sequences. Bot Mar. 54:159–167.
Cho, GY., Boo, SM., Nelson, W. & Clayton, MN. 2005. Genealogical partitioning and phylogeography of Colpomenia peregrina (Scytosiphonaceae, Phaeophyceae), based on plastid rbcL and nuclear ribosomal DNA internal transcribed spacer sequences. Phycologia. 44:103–111.
Cho, GY., Choi, DW., Kim, MS. & Boo, SM. 2009. Sequence repeats enlarge the internal transcribed spacer 1 region of the brown alga Colpomenia sinuosa (Scytosiphonaceae, Phaeophyceae). Phycol Res. 57:242–250.
Clayton, MN. 1975. A study of variation in Australian species of Colpomenia (Phaeophyta, Scytosiphonales). Phycologia. 14:187–195.
Cock, JM., Sterck, L., Rouzé, P., Scornet, D., Allen, AE., Amoutzias, G., Anthouard, V., Artiguenave, F., Aury, J-M., Badger, JH., Beszteri, B., Billiau, K., Bonnet, E., Bothwell, JH., Bowler, C., Boyen, C., Brownlee, C., Carrano, CJ., Charrier, B., Cho, GY., Coelho, SM., Collén, J., Corre, E., Da Silva, C., Delage, L., Delaroque, N., Dittami, SM., Doulbeau, S., Elias, M., Farnham, G., Gachon, CMM., Gschloessl, B., Heesch, S., Jabbari, K., Jubin, C., Kawai, H., Kimura, K., Kloareg, B., Küpper, FC., Lang, D., La Bail, A., Leblanc, C., Lerouge, P., Lohr, M., Lopez, PJ., Martens, C., Maumus, F., Michel, G., Miranda-Saavedra, D., Morales, J., Moreau, H., Motomura, T., Nagasato, C., Napoli, CA., Nelson, DR., Nyvall-Collén, P., Peters, AF., Pommier, C., Potin, P., Poulain, J., Quesneville, H., Read, B., Rensing, SA., Ritter, A., Rousvoal, S., Samanta, M., Samson, G., Schroeder, DC., Ségurens, B., Strittmatter, M., Tonon, T., Tregear, JW., Valentin, K., von Dassow, P., Yamagishi, T., Van de Peer, Y. & Wincker, P. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 465:617–621.
Darriba, D., Taboada, GL., Doallo, R. & Posada, D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772 pp.
Guindon, S. & Gascuel, O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
Guiry, MD. & Guiry, GM. 2019. AlgaeBase, World-wide electronic publication, National University of Ireland, Galway, Available from: http://www.algaebase.org
. Accessed Jan 10, 2019
Guo, X. 1979. Sea level changes since late Pleistocene in China. Sci Geol Sin. 4:330–341.
Hamel, G. 1937. Phaeophyceae de France. Fasc. III. Wolf, Paris, 177–240.
Harper, MA., Cassie, CV., Chang, FH., Nelson, WA. & Broady, PA. 2012. Phylum Ochrophyta: brown and golden-brown algae, diatoms, silicioflagellates, and kin. In : Gordon DP, editor New Zealand Inventory of Biodiversity. Vol 3. Kingdoms Bacteria, Protozoa, Chromista, Plantae, Fungi Canterbury. University Press, Christchurch, 114–163.
Hu, Z-M.., Li, J-J.., Sun, Z-M.., Gao, X., Yao, J-T.., Choi, H-G.., Endo, H. & Duan, D-L. 2017. Hidden diversity and phylogeographic history provide conservation insights for the edible seaweed Sargassum fusiforme in the Northwest Pacific. Evol Appl. 10:366–378.
Hu, Z-M.., Li, J-J.., Sun, Z-M.., Oak, J-H.., Zhang, J., Fresia, P., Grant, WS. & Duan, D-L. 2015. Phylogeographic structure and deep lineage diversification of the red alga Chondrus ocellatus Holmes in the Northwest Pacific. Mol Ecol. 24:5020–5033.
Hu, Z-M.., Zhang, J., Lopez-Bautista, J. & Duan, D-L. 2013. Asymmetric genetic exchange in the brown seaweed Sargassum fusiforme (Phaeophyceae) driven by oceanic currents. Mar Biol. 160:1407–1414.
Keith, SA., Kerswell, AP. & Connolly, SR. 2014. Global diversity of marine macroalgae: environmental conditions explain less variation in the tropics. Glob Ecol Biogeogr. 23:517–529.
Kogame, K. 1997. Life history of Colpomenia sinuosa and Hydroclathrus clathratus (Scytosiphonales, Phaeophyceae) in culture. Phycol Res. 45:277–231.
Kogame, K., Horiguchi, T. & Masuda, M. 1999. Phylogeny of the order Scytosiphonales (Phaeophyceae) based on DNA sequences of rbcL, partial rbcS, and partial LSU nrDNA. Phycologia. 38:496–502.
Kumar, S., Stecher, G. & Tamura, K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
Lee, KM., Boo, GH., Coyer, JA., Nelson, WA., Miller, KA. & Boo, SM. 2014a. Distribution patterns and introduction pathways of the cosmopolitan brown alga Colpomenia peregrina using mt cox 3 and atp 6 sequences. J Appl Phycol. 26:491–504.
Lee, KM., Boo, SM., Kain (Jones), JM. & Sherwood, AR. 2013. Cryptic diversity and biogeography of the widespread brown alga Colpomenia sinuosa (Ectocarpales, Phaeophyceae). Bot Mar. 56:15–25.
Lee, KM., Mansilla, A., Nelson, WA. & Boo, SM. 2012.
Colpomenia durvillei (Scytosiphonaceae, Phaeophyceae): its distribution and relationships with other elongate species of the genus. Bot Mar. 55:367–375.
Lee, KM., Riosmena-Rodriguez, R., Kogame, K. & Boo, SM. 2014b.
Colpomenia wynnei sp. nov. (Scytosiphonaceae, Phaeophyceae): a new species of marine algae from northeast Asia. Phycologia. 53:480–487.
Lee, Y. & Kang, S. 2001. A catalogue of the seaweeds in Korea. Cheju National University Press, Jeju, 662 pp.
Lee, Y. 2008. Marine algae of Jeju. Academy Publication, Seoul, 477 pp.
Li, J-J.., Hu, Z-M.., Gao, X., Sun, Z-M.., Choi, H-G., Duan, D-L.. & Endo, H. 2017. Oceanic currents drove population genetic connectivity of the brown alga Sargassum thunbergii in the north-west Pacific. J Biogeogr. 44:230–242.
Li, T. & Chang, F. 2009. Paleoceanography in the Okinawa Trough. Ocean Press, Beijing, 259 pp.
Librado, P. & Rozas, J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452.
Lin, S-M.., Tseng, L-C.., Ang, PO. Jr, Bolton, J. & Liu, L-C. 2018. Long-term study on seasonal changes in floristic composition and structure of marine macroalgal communities along the coast of Northern Taiwan, southern East China Sea. Mar Biol. 165:83 pp.
Liu, F., Pang, S., Li, J. & Li, X. 2016. Complete mitochondrial genome of the brown alga Colpomenia peregrina (Scytosiphonaceae, Phaeophyceae): genome characterization and comparative analyses. Mitochondrial DNA A DNA Mapp Seq Anal. 27:1601–1603.
Liu, J. 1995. Paleoceanographic characteristics during the LGM period in the west Pacific marginal seas. Mar Sci. 19:14–16.
Liu, R. 2008. Checklist of marine biota of China seas. Science Press, Beijing, 1267 pp.
Longhurst, A. 2007. Ecological geography of the sea. Elsevier Academic Press, Oxford, 560 pp.
Ma, C-L.., Ma, R-P. & Bai, Y-H. 2017. Characteristics of atmospheric environment in China’s coastal areas and atmospheric corrosion in typical coastal regions. Equip Environ Eng. 14:65–69.
Müller, R., Laepple, T., Bartsch, I. & Wiencke, C. 2009. Impact of oceanic warming on the distribution of seaweeds in polar and cold-temperature waters. Bot Mar. 52:617–638.
Nelson, WA. 2013. New Zealand seaweeds: an illustrated guide. Te Papa Press, Wellington, NZ, 328 pp.
Okamura, K. 1936.
Nippon kaisô shi [Descriptions of Japanese algae]. Uchida Rokakuho, Tokyo, 964 pp.
Perestenko, LP. 1980. Vodorosli Zaliva Petra Velikogo [The seaweeds of Peter the Great Bay]. “NAUKA” Leningradskoe Otdelenie, Leningrad, 232 pp.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, DL., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, MA. & Huelsenbeck, JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
Santiañez, WJE., Macaya, EC., Lee, KM., Cho, GY., Boo, SM. & Kogame, K. 2018. Taxonomic reassessment of the Indo-Pacific Scytosiphonaceae (Phaeophyceae): Hydroclathrus rapanuii sp. nov. and Chnoospora minima from Easter Island, with proposal of Dactylosiphon gen. nov. and Pseudochnoospora gen. nov. Bot Mar. 61:47–64.
Sauvageau, C. 1927. Sur le Colpomenia sinuosa Derb. et Sol. Bull Stn Biol d’Arcachon. 24:309–353.
Schaffelke, B., Smith, JE. & Hewitt, CL. 2006. Introduced macroalgae - a growing concern. J Appl Phycol. 18:529–541.
Scott, FJ. 2017. Marine plants of Tasmania. Tasmania Herbarium, Tasmanian Museum and Art Gallery, Hobart, 360 pp.
Stamatakis, A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
Tittensor, DP., Mora, C., Jetz, W., Lotze, HK., Richard, D., Berghe, EV. & Worm, B. 2010. Global patterns and predictions of marine biodiversity across taxa. Nature. 466:1098–1101.
Tseng, CK. 1983. Common seaweeds of China. Science Press, Beijing, 316 pp.
Tseng, CK. 2008. Seaweeds in Yellow Sea and Bohai Sea of China. Science Press, Beijing, 453 pp.
Tseng, CK. & Chang, CF. 1959. On the regional division of the marine algal flora of the western north Pacific. Oceanol Limnol Sin. 2:244–267.
Tseng, CK. & Chang, CF. 1963. A preliminary analytical study of the Chinese marine algal flora. Oceanol Limnol Sin. 5:245–253.
Tyberghein, L., Verbruggen, H., Pauly, K., Troupin, C., Mineur, F. & De Clerck, O. 2012. Bio-ORACLE: a global environmental dataset for marine species distribution modelling. Glob Ecol Biogeogr. 21:272–281.
van den Hoek, C. 1984. World-wide latitudinal and longitudinal seaweed distribution patterns and their possible causes, as illustrated by the distribution of Rhodophytan genera. Helgol Meeresunters. 38:227–257.
Vandermeulen, H. 1984. The taxonomy and autecology of Colpomenia peregrina (Sauv.) Hamel (Phaeophyceae). PhD dissertation. The University of British Columbia, Vancouver, 330 pp.
Wang, P. 1999. Response of western Pacific marginal seas to glacial cycles: paleoceanographic and sedimentological features. Mar Geol. 156:5–39.
Xie, C., Jian, Z. & Zhao, Q. 1996. The paleogeographic configuration of China Seas and its climate influence during the last glacial maximum. Quat Sci. 16:129 pp.
Yoshida, T., Suzuki, M. & Yoshinaga, K. 2015. Checklist of marine algae of Japan (revised in 2015). Jpn J Phycol. 63:129–189.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||