ABSTRACTThe present-day genetic structure of macroalgal species reflects both geographical history and oceanic circulation patterns as well as anthropogenic introduction across native ranges. To precisely understand the genetic diversity and how the factors shape the current population structure of Gloiopeltis furcata sensu lato, we determined the mitochondrial 5′ end of cytochrome c oxidase subunit I (COI-5P) sequences for 677 individuals sampled from 67 sites spanning almost the entire distribution range in Korea and Japan. Results from the phylogenetic analysis and haplotype distribution revealed eleven distinct lineages within G. furcata s.l. along the Korea-Japan coastal areas and displayed divergent phylogeographic patterns among lineages. Despite the closely related lineages distributed in same habitats as high rocky intertidal zone, they display different phylogeographic patterns among lineages. The populations from the south of Korea-Japan harbored the highest genetic diversity and unique endemism in comparison with other populations in the distribution range. This could be the evidence of southern refugia for G. furcata s.l. in the Northwest (NW) Pacific and the recent migration from native to introduced region. The reason is that an exceptional distribution pattern was found high genetic diversity in Hakodate of Japan where is the northern location in the NW Pacific. Our results imply the contemporary influence on the distribution due to current circulation pattern and anthropogenic effects. These phylogeographic findings provide the important insight into cryptic species diversity and the detailed distribution pattern of Gloiopeltis in the NW Pacific.
INTRODUCTIONWithin the past few decades, phylogeographic studies in macroalgae have been increased in the Northwest (NW) Pacific region (Lee et al. 2012, Hu et al. 2016, 2018, Boo et al. 2019, Koh and Kim 2019, Yang et al. 2020). The relatively well-documented geological history of the NW Pacific (such as the sea level fluctuation during the Pleistocene) has facilitated in explaining current genetic patterns observed for different macroalgal species (Yang et al. 2009, Cheang et al. 2010, Hu et al. 2011, 2015). The considerable climatic changes experienced across Quaternary glacial-interglacial cycles have left marked genetic signatures in the current geographical distribution of marine macroalgae (Provan and Bennett 2008, Hu et al. 2018). Advancing glacial ice caused the sea level to fall 120–130 m, consequently, sea floor exposed (Wang 1999). During these sudden changes in the environment, marine temperate species had retreated southward as refugia with stable and diverse conditions for survival, facilitating persistence with regional biotic and abiotic environmental changes (Hewitt 1996, Neiva et al. 2015). In populations in such refugia have been functioned as initial genetic resources for the formation of distinct evolutionary lineages, and species have colonized new regions from environmental conditions improved (Neiva et al. 2015). Through these processes, it is generally recognized that lower-latitude regions host populations with higher genetic diversity than those in areas where have been recently recolonized (Neiva et al. 2015).
Environmental factors, including current circulation pattern, also differentially affect the distribution pattern of seaweeds (Li et al. 2017, Boo et al. 2019). Complex oceanic circulation in the NW Pacific influences the regional population genetic structure and biogeographic history of macroalgae with limited dispersal abilities (Hu et al. 2015). For example, the north-flowing Kuroshio Current divides into its branches, the Tsushima Warm Current and the Yellow Sea Warm Current (Hu et al. 2015). The Tsushima Warm Current (TWC1) runs northward and branched into the Tsugaru Warm Current (TWC2), which flow through the Tsugaru Strait and then southward along the coast of the north-east of the main island of Japan (Li et al. 2017).
Apart from these historical and environmental processes, the anthropogenic transport of species from native ranges to new areas has become commonplace, resulting in processes responsible for the present-day population distribution and genetic diversity in macroalgal species (Andreakis et al. 2016, Díaz-Tapia et al. 2018). Actually, a variety of macroalgae have been reported as invasive species by human activities and they have probably been introduced as microscopic life stages or resting forms settled either on shellfish or on packing material (Kim et al. 2010, Cecere et al. 2011). Shipping is main vector of long-range transport of macroalgae because invasive haplotype occurred in harbors or near harbors (Kim et al. 2010, Geoffroy et al. 2016).
The red macroalga Gloiopeltis is distributed in the North Pacific from the Philippines in the south to Alaska in the north, forming extensive populations in high intertidal rocky zones (Oh and Lee 1996, Guiry and Guiry 2020). Molecular studies of Gloiopeltis have provided new insights into taxonomy, biodiversity and phylogeography (Yang and Kim 2018, Hanyuda et al. 2019, 2020, Yang et al. 2020). Gloiopeltis furcata sensu lato (s.l.) have wide distribution range among the Gloiopeltis species and distinguished by its hollow and cylindrical to subcylindrial thalli (Lee et al. 1996, Guiry and Guiry 2020). It was recently shown to be composed of morphologically similar but genetically distinct lineages based on mitochondrial and plastid genes (Hanyuda et al. 2019, Yang et al. 2020). The phylogeographic study of G. furcata s.l. from the North Pacific revealed the evidence of eight distinct lineages, and the diversification of this group occurred mostly within the NW Pacific (Yang et al. 2020). Meanwhile, only G. furcata sensu stricto (s.s.) is widely distributed along both sides of the North Pacific, ranging from Korea, Japan, and Kamchatka to Alaska, British Columbia, and Washington (Hanyuda et al. 2019, Yang et al. 2020). Despite their studies, the detailed distribution pattern of G. furcata s.l. within the NW Pacific is not fully understood because of insufficient collections across the distribution range of G. furcata s.l. in Korea and Japan.
To improve our understanding of phylogeography within the NW Pacific, we conducted extensive sampling of G. furcata s.l. across most of its geographical range in Korea and Japan and used the 5′ end of cytochrome c oxidase subunit I (COI-5P) marker to elucidate its genetic diversity, phylogenetic relationships and phylogeographic patterns. Mitochondrial COI-5P has proven the value for phylogeographic study of G. furcata s.l., showing high genetic variation (Yang et al. 2020). In the current study, molecular phylogeny was used to clarify the species diversity of G. furcata s.l. Distribution patterns within the NW Pacific were explored by evaluating the genetic structure of populations. Finally, we tested whether a glacial refugia existed around the southern region in the NW Pacific, which may have contributed to diversity resources.
MATERIALS AND METHODSSamples of G. furcata s.l. were collected from 67 sites along the coasts of Korea and Japan (Table 1). Sample were divided into eleven regions based on the criteria by Yang et al. (2020) in Korea and local boundary in Japan (West coast of Korea [WEK], Jeju in Korea [JJK], South coast of Korea [SOK], East coast of Korea [EAK], Kyushu of Japan [KYS], Shikoku of Japan [SKK], Kansai of Japan [KAS], Shikinejima of Japan [SKJ], Kanto of Japan [KAT], Tohoku of Japan [THK], Hokkaido of Japan [HOK]). On the East Sea (Sea of Japan) coasts, Gloiopeltis is absent or quite rare because of the small tide (Saito 1970). All specimens studied herein were deposited in the herbarium of Jeju National University (JNUB). Epiphytes on specimens were removed under a dissecting microscope before DNA extraction.
Apical regions were separated from the thallus, cleaned and stored in silica gel for molecular analyses. DNA was extracted using the LaboPass Tissue Genomic DNA Isolation Kit (Cosmogenetech, Seoul, Korea), according to the manufacturer’s protocol. Targeted gene sequences of COI-5P were amplified and sequenced using the primers GWSFn–GWSRx (Le Gall and Saunders 2010, Saunders and McDevit 2012). All polymerase chain reaction (PCR) amplifications were carried out in All-In-One-Cycler (Bioneer, Daejeon, Korea) using MasterMix 2× (MGmed, Seoul, Korea), under the following conditions: 4 min of initial denaturation at 96°C, and 40 cycles of 30 s for denaturation, 30 s at 45°C for annealing, and 1 min at 72°C for extension, with a final extension at 72°C for 7 min. All sets of PCR included a negative control reaction tube containing all reagents, except template DNA. The PCR products were purified using Exo-AP PCR Clean-up Mix (MGmed) and then sequenced commercially (Macrogen, Seoul, Korea).
Sequences of the forward and reverse strands were determined for all specimens, and the electropherograms were edited using the program Chromas v.1.45 (McCarthy 1998) and checked manually for consistency. Consensus sequences were generated using Geneious software (Geneious R9 ver. 9.1.4, http://www.geneious.com). Sixty-five sequences from Korea and Japan, previously analyzed by Yang et al. (2020), were downloaded from GenBank and aligned with newly generated sequences in this study. Sequences were aligned manually using Clustal O (Sievers et al. 2011). Unique haplotype sequences of COI-5P yielded in this study were deposited in GenBank (MW271526–MW271613) (Supplementary Table S1).
Phylogenetic reconstruction of the COI-5P gene was conducted using the maximum likelihood (ML) method using RAxML v8 (Stamatakis 2006) with 1,000 bootstrap replicates, based on the GTRGAMMA model. Twenty-eight sequences of four Gloiopeltis species, G. tenax, G. complanata, G. frutex, and G. compressus, from GenBank were also downloaded and included in the alignments for phylogenetic analysis. Endocladia muricata (KF026475, KM254659, and KM254306) was included as outgroup based on previous results (Yang and Kim 2018).
DNA polymorphism levels of COI-5P were summarized in terms of number of haplotypes (Nh), haplotype diversity (h), and nucleotide diversity (π) using Arlequin v. 3.5.2 (Excoffier et al. 2005). To evaluate the relationships among haplotypes, we constructed a haplotype network with a minimum spanning network in Arlequin. Differences in the genetic structure of populations were assessed by computing pairwise FST statistics in Arlequin v. 3.5 between locations. The Kimura 2-parameter model (Kimura 1980) was used to calculate pair-wise distances within and between lineages. To neutrality tests, the Tajima’s D (Tajima 1989) and Fu’s FS (Fu 1997) implemented in Arlequin were used to test for population expansion.
RESULTSWe obtained a total of 677 COI-5P sequences of G. furcata s.l., of which 476 were newly generated in this study. The COI-5P alignment was composed of 567 bp and yielded 153 haplotypes (Table 2). The phylogeny inferred from these haplotypes with other sequences of Gloiopeltis from GenBank showed fifteen genetically distinct lineages within the genus Gloiopeltis, including four previously described species, G. tenax, G. complanata, G. frutex, and G. compressus (Fig. 1A). Thirteen of these lineages were moderately to highly supported lineages (bootstrap values >68 for ML) while the remaining two, lineage 2 and lineage 10, were singletons. The ML phylogenetic tree suggested the presence of eleven putative species (G. furcata s.s. and lineage 1–10) within the G. furcata s.l. group. Among them, three lineages were newly discovered in this study and eight were already observed in previous study by Yang et al. (2020). The clade of G. furcata s.l. include G. tenax and G. complanata, which were sistered with lineage 2 and lineage 10, respectively.
Mapping the distribution of eleven lineages of G. furcata s.l. along the coasts of Korea and Japan showed that they differ distribution patterns (Fig. 1B). Among the lineages, seven lineages co-distributed in Korea and Japan. First of all, G. furcata s.s. (consisting of 25 haplotypes) was only distributed in cold water coastal areas, such as the east of Korea (EAK) and the north of Japan (HOK and THK). Lineage 1, consisting of 35 haplotypes, was mainly distributed in the south of Japan (KYS, SKK, and KAS) and partly distributed in JJK of Korea and THK of Japan. Lineage 3, consisting of 22 haplotypes, was mainly distributed in the south of Korea / Japan, as well as an unusual disjunct occurrence was detected in Hakodate. The widely distributed lineage 5, consisting of 36 haplotypes, was found in the south of Korea (JJK and SOK), and the south and north of Japan (KYU, SKK, THK, and HOK). Lineage 6, consisting of nine haplotypes, was mainly distributed in Korea (WEK, SOK, and JJK) with a disjunct occurrence in Hakodate. Lineage 7, consisting of eight haplotypes, was distributed in Korea (WEK, SOK, and JJK) as well as in Japan (KYS, THK, and HOK). Lineage 9, consisting of three haplotypes, was distributed in the south of Korea / Japan (JJK, KYS, and SKK). Four lineages only occurred in Japan. Lineage 8, consisting of ten haplotypes, was distributed in Japan from the south to north (KYS, SKK, KAT, THK, and HOK). Lineage 4, consisting of four haplotypes, was only distributed in one population in SKJ of Japan. Lineages 2 and 10 were only found in KYS of Japan. Most phylogenetically diverged lineages occurred in the southern region, in KYS of Japan and JJK of Korea, and an exceptional pattern appeared in Hakodate of HOK, Japan.
Molecular diversity indices showed that the populations from Korea (JJK and SOK) and Japan (KYS, SKK, and HOK) had high genetic diversity (h = 0.8558–0.9516, π = 0.0769–0.1068), whereas the populations from SKJ, KAT, and THK in Japan had lower values (h = 0.3846–0.6952, π = 0.0016–0.0441) (Table 3). The highest value of diversity was detected in KYS of Japan (h = 0.9516, π = 0.1013), which also contained the largest number of lineages (n = 8). KAS of Japan and EAK of Korea had relatively high haplotype diversity (0.8832–0.9044), but the nucleotide diversity was low (0.0037–0.0541). For the lineages of G. furcata s.l., Tajima’s D and Fu’s FS values were negative in three lineages (Table 2), indicating that population of lineages 1, 6, and 8 expanded in the past.
Pairwise FST values based on COI-5P data (Table 4) revealed low to moderate values within Korea (FST range = 0.040–0.456). The populations from Korea showed genetic affinity with those from the southern region of Japan (KYS, SKK; FST range = 0.2128–0.3492), and quite close to those from HOK of Japan (FST range = 0.3343–0.4057). Moderate to high FST values were detected between populations from Korea (WEK, JJK, SOK, and EAK) and the central region of Japan (KAS, SKJ, KAT, and THK: FST range = 0.4849–0.8975). In contrast, FST values showed large genetic variance within populations in Japan (FST range = 0.0861–0.9867). The population from KAS and SKJ of Japan was significantly divergent from that of other populations (KAS: FST range = 0.3859–0.9756; SKJ: FST range = 0.3764–0.9867).
The COI-5P haplotype network yielded several unconnected haplotype groups corresponding to the nine different phylogenetic lineages, except singletons (lineages 2 and 10) (Fig. 2). Among all haplotypes (153), 25 were shared between sampling sites in Korea and Japan, and 128 haplotypes were site-specific. Within six lineages (G. furcata s.s. and lineages 1, 3, 5, 7, and 9), many missing haplotypes over 10 steps were detected (Fig. 2). In G. furcata s.s., haplotypes fell into three highly divergent and geographically distinct clusters: West HOK of Japan, South / East HOK of Japan, and EAK of Korea. Likewise, lineage 1 were separated into four highly divergent clusters, which roughly matched their geographical distribution: KAS, SKK / KYS, and THK. In lineage 5, haplotypes were separated into several clusters and exhibited a clear phylogeographic structure between the West HOK and South HOK of Japan. On the other hand, lineages 3, 6, and 7 showed that the haplotypes from Korea (WEK, JJK, and SOK) and Japan (KYS and SKK) tended to be related to each other, resulting from shared and less differentiated haplotypes. Haplotypes of the lineages 4, 8, and 9 occurred in Japan only. In lineage 9, haplotypes were separated into two geographical clusters, KYS and SKK, by 26 missing steps despite close region to each other.
DISCUSSIONCurrently, five Gloiopeltis species have been accepted from the NW Pacific region: G. tenax, G. complanata, G. furcata, G. frutex, and G. compressus (Yang and Kim 2018, Hanyuda et al. 2019, 2020, Guiry and Guiry 2020). Our results provide the evidence of eleven different lineages within G. furcata s.l. along the Korea-Japan coastal areas that may be considered to be distinct species. Our new sequences mostly assign to previously observed lineage by Yang et al. (2020), meanwhile, some are presented as newly detected lineage. In summarizing previously described species and newly detected lineage in this study, at least fifteen lineages of Gloiopeltis may exist in the NW Pacific region. This suggests that high levels of cryptic species diversity have been still undiscovered in macroalgal groups, especially in the NW Pacific (Hu et al. 2015, Yang and Kim 2018, Boo et al. 2019, Lee et al. 2020). Our results reveal the phylogeographic pattern of G. furcata s.l. throughout the Korea-Japan region in the NW Pacific. Most phylogeographic studies on macroalgae have focused on one or two species within the genus to illustrate distribution patterns in the NW Pacific (Hu et al. 2011, Kim et al. 2012, Lee et al. 2012, Hu et al. 2018, Boo et al. 2019). Our study on G. furcata s.l., including 11 co-distributed distinct lineages in high intertidal zone, provides important insight into the comprehensive biogeography of Gloiopeltis in this region. The haplotype network based on COI-5P facilitates a better understanding of their demographic histories.
Molecular evidence suggests the existence of a southern glacial refugia for G. furcata s.l. in the NW Pacific. Genetic signatures of glacial refugia typically include a high level of genetic diversity and endemism of marine organisms (Hewitt 2004, Provan et al. 2005, Hu et al. 2018). Our results reveal that the highest level of haplotype and nucleotide diversity were found mostly in the south of Korea (JJK and SOK) and Japan (KYS). Moreover, most diverse lineages of G. furcata s.l. occurre in JJK of Korea and KYS of Japan. Several of them are endemic because they have not been detected elsewhere in middle-to-northern Korea and Japan. Such distribution patterns are in line with the hypothesis that temperate populations in the northern hemisphere could survive with a high genetic diversity in the south refugia, because the majority of species retreated to the south in the northern hemisphere following climate changes (Hewitt 2004).
The identification of glacial refugia in the NW Pacific has been documented in several marine macroalgae (Cheang et al. 2010, Hu et al. 2011, Kim et al. 2012, Lee et al. 2012, Hu et al. 2016, Boo et al. 2019). Similar to our results of G. furcata s.l., the southern refugia has been found in the NW Pacific in the following species: Ishige okamurae (southern Korea including Jeju Island) (Lee et al. 2012), Gelidiophycus freshwateri (southern Korea) (Boo et al. 2019), Gelidium elegans (Jeju and central Pacific Japan) (Kim et al. 2012), and Sargassum horneri (Kyushu) (Hu et al. 2011). These findings are also consistent with the existence of southern refugia in the north Atlantic for marine macroalgae (for example, Palmaria palmata, Provan et al. 2005; Fucus serratus, Hoarau et al. 2007; and Chondrus crispus, Hu et al. 2011). The southern coasts of Korea-Japan might provide suitable habitats during glacial maxima for intertidal algal species, which frequently accord to the results of rear edge distribution showing high genetic diversity and / or endemism in low-latitude refugia in the Northwest Atlantic (Neiva et al. 2020). The low-latitude region at the rear edge is regarded as an important area for genetic diversity hotspots and should thus receive special conservation efforts due to its unique genetic diversity (Provan et al. 2005).
An exceptional distribution pattern is found in HOK of Japan, demonstrating high genetic diversity despite its northern location in the NW Pacific. We discover the same haplotypes in HOK of Japan and south of Korea-Japan (H02 of lineage 8 and H67 of lineage 7), which occurred in Hakodate, Muroran and Shiragami. Less differentiated haplotypes (H70–H127 of lineage 3 and H69–H104 of lineage 6) also found between HOK and the south of Korea-Japan. It suggests that populations could have been introduced to HOK by anthropogenic transport. High levels of genetic affinity observed between HOK and the south of Korea-Japan indicate the possible occurrence of anthropogenic transportation of genotypes across native ranges. The human-mediated movement of populations from native to introduced regions causes rapid expansion of population distribution and alter natural phylogeographic patterns (Straub et al. 2016). Anthropogenically altered distribution patterns have often been demonstrated in macroalgae with low dispersal capabilities, such as Asparagopsis taxiformis, Symphyocladia dendroidea, and Polysiphonia devoniensis (Andreakis et al. 2016, Díaz-Tapia et al. 2018). Even within HOK, distinct lineages of G. furcata s.l. that were not found in the proximity are predominant in Hakodate (Fig. 1B). The port of Hakodate, where is near the site with high genetic diversity of G. furcata s.l., has been opened since 1854 as one of the first international trading port in Japan (Ivings 2017). Alternatively, genetic connectivity between Hakodate and southern Korea-Japan could also be explained by the Tsushima Warm Current (Li et al. 2017). In fact, it has been reported that a mass of drifting algae including Gloiopeltis appeared to have originated in the coast of KYS of Japan, passed through the Sado Strait with the northward flow of the Tsushima Warm Current (Ikehara and Sano 1986). As G. furcata s.l. has hollow thallus (Lee et al. 1996), it can be transported by floating. However, the distribution pattern is difficult to explain by the Tsushima Warm Current alone, because the genetic diversity in the west of HOK should be at least equal or similar than the genetic diversity of G. furcata s.l. in Hakodate. Therefore, we assume that the current population distribution in Hakodate has been influenced by combined factors of anthropogenic activities and current circulation pattern in this region.
High levels of genetic divergence among the lineages of G. furcata s.l. suggest long-term isolation of populations (Fig. 2), allowing for independent development of distinct mitochondrial lineages (Pauls et al. 2006). In addition, there was evidence of geographically isolated populations, indicating a poor self-dispersal ability of this algae. Deep genetic divergences within G. furcata s.s. (25 missing steps) indicate that the western HOK of Japan has completely blocked gene flow to the south / east HOK by probably current circulation pattern. This genetic discontinuity is also detected in the population of lineage 5, demonstrating a significant barrier to gene flow between the west and south of HOK (Fig. 2). The populations distributed in THK of Japan in this study have gene exchanges with the population from the southern HOK. Contrary to our results, other studies of macroalgae have revealed the genetic exchange among populations in HOK with low genetic differentiation (Zhang et al. 2015). To understand the population distribution pattern in HOK of Japan, it will be necessary to carry out additional phylogeographic studies, including a better representation of different species distributed in similar habitats to G. furcata s.l.
Seawater temperature is one of the main factors determining the geographical distribution of red algae in the NW Pacific (Boo et al. 2019). For example, Gelidiophycus freshwateri and G. divaricatus adapted to different seawater temperate condition, which is influenced to current species range (Boo et al. 2019). Like this, the distribution range of the lineages of G. furcata s.l. may reflect an adaptation of the species associated with seawater temperature. Despite the similar habitat of G. furcata s.l., they display different geographic distribution ranges based on the COI-5P gene. Gloiopeltis furcata s.s. is distributed in the temperate to cold water region, EAK of Korea and HOK, THK of Japan (Hanyuda et al. 2019, Yang et al. 2020). Gloiopeltis furcata s.s. is the only species of the genus known to be distributed in the East Pacific (Yang et al. 2020). According to a study on the effects of light and temperature on Gloiopeltis development (Chen et al. 2011), the different distribution range of G. tenax and G. furcata are related to the adaptation of low water temperatures. The growth, sprouting and survival of G. furcata discs prefer lower temperatures than G. tenax. Consequently G. furcata could be distributed to the north. Similarly, temperature effect on Gloiopeltis development would have determined the distribution range for each lineage of G. furcata s.l.
In conclusion, we present important insights into Gloiopeltis diversity and phylogeography in the NW Pacific. It is a remarkable example of how closely related species within the genus with habitat specialization as high rocky intertidal zone display divergent phylogeographic patterns. We evidence the southern refugia for G. furcata s.l. in the NW Pacific based on the molecular phylogeography. Our data illustrate not only the natural phylogeographic patterns but also the anthropogenic influence on the distribution, evidenced in Hakodate of Japan. Since Gloiopeltis is one of the most important commercial seaweed in the NW Pacific, taxonomic studies should be conducted to describe new species with an emphasis on morphological observation. In addition, future research is needed to expand the survey areas from south to the north in the NW Pacific to investigate the genetic structure and historical demography of Gloiopeltis populations in this region.
ACKNOWLEDGEMENTSThis research was supported by the 2020 scientific promotion funded by Jeju National University to M. S. Kim, and the Basic Science Research Program (2019R1A6A1A10072987 and 2020R1I1A2069706) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education of Korea.
SUPPLEMENTARY MATERIALS
Supplementary Table S1. Distribution of mtDNA haplotypes (H) and GenBank accession numbers of Gloiopeltis furcata sensu lato (https://www.e-algae.org). CONFLICTS OF INTERESTCONFLICTS OF INTEREST The authors declare that they have no potential conflicts of interest. Fig. 1Results of the molecular analysis of Gloiopeltis furcata s.l. collected from the Korea-Japan coast. (A) Maximum likelihood phylogenetic tree of 5′ end of cytochrome c oxidase subunit I (COI-5P) of G. furcata s.l. including four currently recognized Gloiopeltis species. The outgroup for the analysis was three sequences of Endocladia muricata. Mitochondrial lineages have been labeled with different colors. Only bootstrap support values >50 are indicated in the main nodes. (B) Map showing sampling sites and geographical distribution of the mitochondrial lineages of G. furcata s.l. Circles represent the mitochondrial lineage diversity within each site and circle size is proportional to the number of individuals. Colors correspond to the lineages indicated in the phylogenetic tree (A). Number in circle corresponds to the sampling site in Table 1. 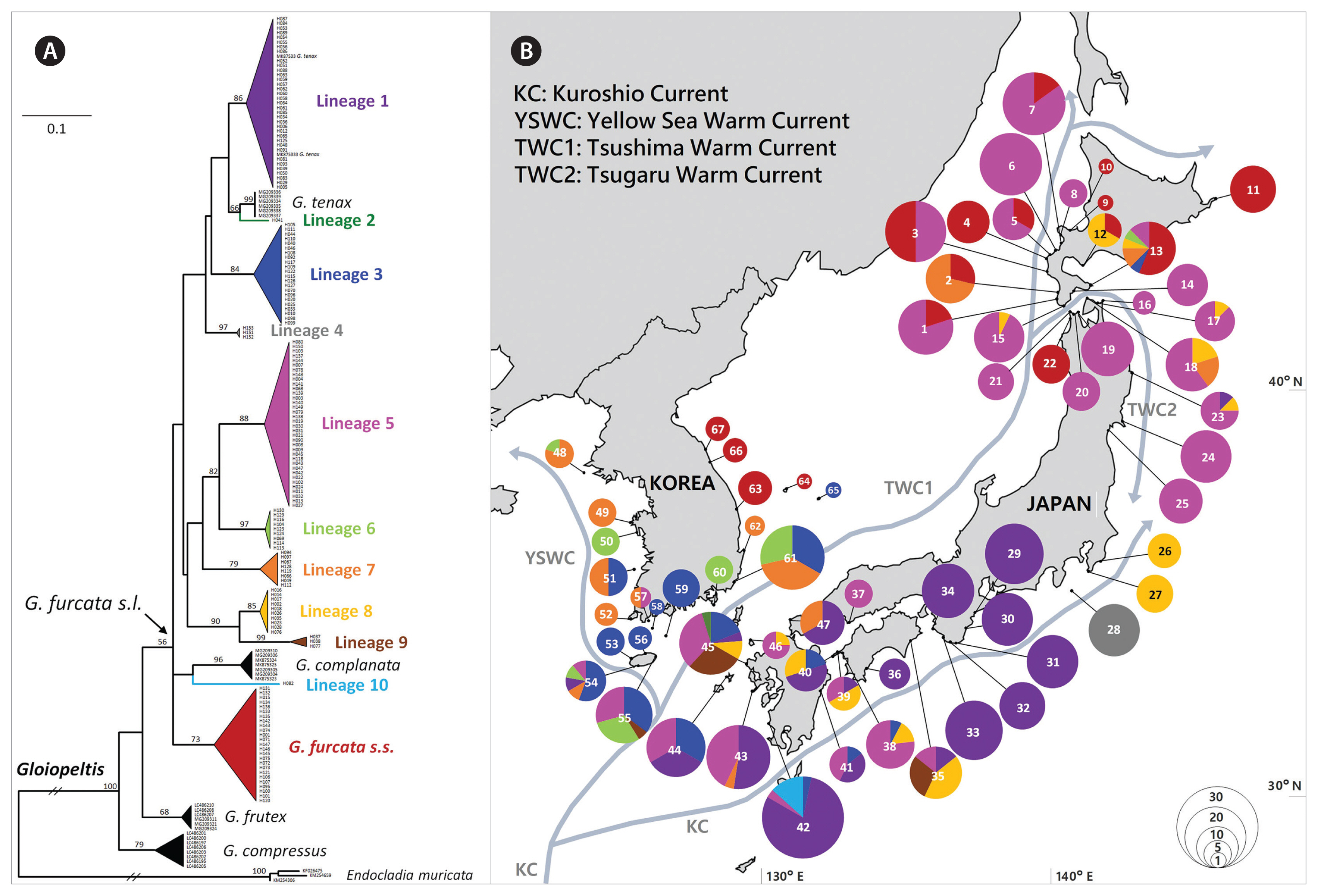
Fig. 2Sampling regions (upper left) and 5′ end of cytochrome c oxidase subunit I (COI-5P) haplotype networks for nine lineages of Gloiopeltis furcata s.l. Circles represent haplotypes and their size is proportional to the number of individuals per haplotype. Colors of circles indicate locations in which haplotypes were found. Each connecting line indicates one mutation step between haplotypes. The vertical bars represent the number of mutational steps between two haplotypes when >1. WEK, West coast of Korea; JJK, Jeju in Korea; SOK, South coast of Korea; EAK, East coast of Korea; KYS, Kyushu of Japan; SKK, Shikoku of Japan; KAS, Kansai of Japan; SKJ, Shikinejima of Japan; KAT, Kanto of Japan; THK, Tohoku of Japan; HOK, Hokkaido of Japan. 
Table 1Sampling information and lineages for Gloiopeltis furcata s.l. included in this study Table 2Summary of molecular diversity and demographic analyses for 11 lineages of Gloiopeltis furcata s.l. from Korea and Japan Table 3Genetic diversity of populations of Gloiopeltis furcata s.l. from Korea and Japan based on COI-5P data COI-5P, 5′ end of cytochrome c oxidase subunit I; N, number of analyzed samples; Nh, number of haplotypes; h, haplotype diversity; π, nucleotide diversity; SD, standard deviation; WEK, West coast of Korea; JJK, Jeju in Korea; SOK, South coast of Korea; EAK, East coast of Korea; KYS, Kyushu of Japan; SKK, Shikoku of Japan; KAS, Kansai of Japan; SKJ, Shikinejima of Japan; KAT, Kanto of Japan; THK, Tohoku of Japan; HOK, Hokkaido of Japan. Table 4Pairwise estimates of FST between populations of Gloiopetlis furcata s.l. based on COI-5P data COI-5P, 5′ end of cytochrome c oxidase subunit I; WEK, West coast of Korea; JJK, Jeju in Korea; SOK, South coast of Korea; EAK, East coast of Korea; KYS, Kyushu of Japan; SKK, Shikoku of Japan; KAS, Kansai of Japan; SKJ, Shikinejima of Japan; KAT, Kanto of Japan; THK, Tohoku of Japan; HOK, Hokkaido of Japan. Significant p-values are indicated by *p < 0.05 and **p < 0.01. REFERENCESAndreakis, N., Costello, P., Zanolla, M., Saunders, GW. & Mata, L. 2016. Endemic or introduced? Phylogeography of Asparagopsis (Florideophyceae) in Australia reveals multiple introductions and a new mitochondrial lineage. J Phycol. 52:141–147.
Boo, GH., Qiu, Y-X., Kim, JY., Ang, PO. Jr, Bosch, S., De Clerck, O., He, P., Higa, A., Huang, B., Kogame, K., Liu, S-L., van Nguyen, T., Suda, S., Terada, R., Miller, KA. & Boo, SM. 2019. Contrasting patterns of genetic structure and phylogeography in the marine agarophytes Gelidiophycus divaricatus and G. freshwateri (Gelidiales, Rhodophyta) from East Sea. J Phycol. 55:1319–1334.
Cecere, E., Moro, U., Wolf, MA., Petrocelli, A., Verlaque, M. & Sfriso, A. 2011. The introduced seaweed Grateloupia turuturu (Rhodophyta, Halymeniales) in two Mediterranean trasitional water systems. Bot Mar. 54:23–33.
Cheang, CC., Chu, KH. & Ang, PO. Jr 2010. Phylogeography of the marine macroalga Sargassum hemiphyllum (Phaeophyceae, Heterokontophyta) in northwestern Pacific. Mol Ecol. 19:2933–2948.
Chen, S., Wu, J., Chen, L. & Zhu, C. 2011. Effects of light and temperature on the attachment and development of Gloiopeltis tenax and Gloiopeltis furcata tetraspores. J Appl Phycol. 23:1045–1051.
Díaz-Tapia, P., Maggs, CA., Macaya, EC. & Verbruggen, H. 2018. Widely distributed red algae often represent hidden introductions, complexes of cryptic species or species with strong phylogeographic structure. J Phycol. 54:829–839.
Excoffier, L., Laval, G. & Schneider, S. 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 1:47–50.
Fu, YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 147:915–925.
Geoffroy, A., Destombe, C., Kim, B., Mauger, S., Raffo, MP., Kim, MS. & Le Gall, L. 2016. Patterns of genetic diversity of the cryptogenic red alga Polysiphonia morrowii (Ceramiales, Rhodophyta) suggest multiple origins of the Atlantic populations. Ecol Evol. 6:5635–5647.
Guiry, MD. & Guiry, GM. 2020. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. Available from: http://www.algaebase.org
. Accessed Aug. 5:2020 pp.
Hanyuda, T., Yamamura, K., Boo, GH., Miller, KA., Vinogradova, KL. & Kawai, H. 2019. Identification of true Gloiopeltis furcata (Gigartinales, Rhodophyta) and preliminary analysis of its biogeography. Phycol Res. 68:161–168.
Hanyuda, T., Yamamura, K. & Kawai, H. 2020. Molecular studies of Gloiopeltis (Endocladiaceae, Gigartinales), with recognition of G. compressus comb. nov. from Japan. Phycologia. 59:1–5.
Hewitt, GM. 1996. Some genetic consequences of ice ages, and their role, in divergence and speciation. Biol J Linn Soc. 58:247–276.
Hewitt, GM. 2004. Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B Biol Sci. 359:183–195.
Hoarau, G., Coyer, JA., Veldsink, JH., Stam, WT. & Olsen, JL. 2007. Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus
. Mol Ecol. 16:3606–3616.
Hu, Z-M., Kantachumpoo, A., Liu, R-Y., Sun, Z-M., Yao, J-T., Komatsu, T., Uwai, S. & Duan, D-L. 2018. A late Pleistocene marine glacial refugium in the south-west of Hainan Island, China: phylogeographic insights from the brown alga Sargassum polycystum
. J Biogeogr. 45:355–366.
Hu, Z-M., Li, J-J., Sun, Z-M., Gao, X., Yao, J-T., Choi, H-G., Endo, H. & Duan, D-L. 2016. Hidden diversity and phylogeographic history provide conservation insights for the edible seaweed Sargassum fusiforme in the Northwest Pacific. Evol Appl. 10:366–378.
Hu, Z-M., Li, J-J., Sun, Z-M., Oak, J-H., Zhang, J., Fresia, P., Grant, WS. & Duan, D-L. 2015. Phylogeographic structure and deep lineage diversification of the red alga Chondrus ocellatus Holmes in the Northwest Pacific. Mol Ecol. 24:5020–5033.
Hu, Z-M., Uwai, S., Yu, S-H., Komatsu, T., Ajisaka, T. & Duan, D-L. 2011. Phylogeographic heterogeneity of the brown macroalga Sargassum horneri (Fucaceae) in the northwestern Pacific in relation to late Pleistocene glaciation and tectonic configurations. Mol Ecol. 20:3894–3909.
Ikehara, K. & Sano, O. 1986. Distribution and species composition of floating seaweeds collected in the Sado Straits of the Japan Sea. Bull Jpn Sea Reg Fish Res Lab. 36:59–75.
Ivings, S. 2017. Trade and Conflict at the Japanese Frontier: Hakodate as a Treaty Port, 1854–1884. J Transcult Stud. 8:103–137.
Kim, KM., Hoarau, GG. & Boo, SM. 2012. Genetic structure and distribution of Gelidium elegans (Gelidiales, Rhodophyta) in Korea based on mitochondrial cox 1 sequence data. Aquat Bot. 98:27–33.
Kim, SY., Weinberger, F. & Boo, SM. 2010. Genetic data hint at a common donor region for invasive Atlantic and Pacific populations of Gracilaria vermiculophylla (Gracilariales, Rhodophyta). J Phycol. 46:1346–1349.
Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Ecol. 16:111–120.
Koh, YH. & Kim, MS. 2019. Genetic diversity and distribution pattern of economic seaweeds Pyropia yezoensis and Py. suborbiculata (Bangiales, Rhodophyta) in the northwest Pacific. J Appl Phycol. 32:2495–2504.
Le Gall, L. & Saunders, GW. 2010. DNA barcoding is a powerful tool to uncover algal diversity: a case study of the Phyllophoraceae (Gigartinales, Rhodophyta) in the Canadian flora. J Phycol. 46:374–389.
Lee, HW., Bae, EH. & Kim, MS. 2020.
Umbraulva yunsella sp. nov. (Ulvaceae, Chlorophyta) from a subtidal habitat of Jeju Island, Korea. Algae. 35:349–359.
Lee, JW., Oh, B-G. & Lee, H-B. 1996. Morphological variations of Gloiopeltis furcata (Postels et Ruprecht) J. Agardh (Rhodophyta) in the East coast of Korea. Algae. 11:91–94.
Lee, KM., Yang, EC., Coyer, JA., Zuccarello, GC., Wang, W-L., Choi, CG. & Boo, SM. 2012. Phylogeography of the seaweed Ishige okamurae (Phaeophyceae): evidence for glacial refugia in the northwest Pacific region. Mar Biol. 159:1021–1028.
Li, J-J., Hu, Z-M., Gao, X., Sun, Z-M., Choi, H-G., Duan, D-L. & Endo, H. 2017. Oceanic currents drove population genetic connectivity of the brown alga Sargassum thunbergii in the north-west Pacific. J Biogeogr. 44:230–242.
McCarthy, C. 1998. Chromas version 1.45. School of Health Science, Griffith University, Southport..
Neiva, J., Assis, J., Coelho, NC., Femandes, F., Pearson, GA. & Serrão, EA. 2015. Genes left behind: climate change threatens cryptic genetic diversity in the canopy-forming seaweed Bifurcaria bifurcata
. PLoS ONE. 10:e0131530 pp.
Neiva, J., Serrão, EA., Paulino, C., Gouveia, L., Want, A., Tamigneaus, É., Ballenghien, M., Mauger, S., Fouqueau, L., Engel-Gautier, C., Destombe, C. & Valero, M. 2020. Genetic structure of amphi-Atlantic Laminaria digitata (Laminariales, Phaeophyceae) reveals a unique range-edge gene pool and suggests post-glacial colonization of the NW Atlantic. Eur J Phycol. 55:517–528.
Oh, B-G. & Lee, H-B. 1996. Morphology of three species of Gloiopeltis (Endocladiaceae, Rhodophyta) in Korea. Algae. 11:81–90.
Pauls, SY., Thorsten Lumbsch, H. & Haase, P. 2006. Phylogeography of the montane caddisfly Drusus discolor: evidence for multiple refugia and periglacial survival. Mol Ecol. 15:2153–2169.
Provan, J. & Bennett, KD. 2008. Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol. 23:564–571.
Provan, J., Wattier, RA. & Maggs, CA. 2005. Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Mol Ecol. 14:793–803.
Saito, Y. 1970. The reason of poor macroalgal flora along the coast of Sea of Japan: from an ecological point of view. Kagaku. 40:561–565.
Saunders, GW. & McDevit, DC. 2012. Acquiring DNA sequence data from dried archival red algae (Florideophyceae) for the purpose of applying available names to contemporary genetic species: a critical assessment. Botany. 90:191–203.
Sievers, F., Wilm, A., Dineen, D., Gibson, TJ., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., Söding, J., Thompson, JD. & Higgins, DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539 pp.
Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
Straub, SC., Thomsen, MS. & Wernberg, T. 2016. The dynamic biogeography of the anthropocene: the speed of recent range shifts in seaweeds. In : Hu Z-M, Fraser C, editors Seaweed Phylogeography. Springer, Amsterdam, 63–93.
Tajima, F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 123:585–595.
Wang, P. 1999. Response of Western Pacific marginal seas to glacial cycles: paleoceanographic and sedimentological features. Mar Geol. 156:5–39.
Yang, EC., Lee, SY., Lee, WJ. & Boo, SM. 2009. Molecular evidence for recolonization of Ceramium japonicum (Ceramiaceae, Rhodophyta) on the west coast of Korea after the last glacial maximum. Bot Mar. 52:307–315.
Yang, MY. & Kim, MS. 2018. DNA barcoding of the funoran-producing red algal genus Gloiopeltis (Gigartinales) and description of a new species, Gloiopeltis frutex sp. nov. J Appl Phycol. 30:1381–1392.
Yang, MY., Yang, EC. & Kim, MS. 2020. Genetic diversity hotspot of the amphi-Pacific macroalga Gloiopeltis furcata sensu lato (Gigartinales, Florideophyceae). J Appl Phycol. 32:2515–2522.
Zhang, J., Yao, J-T., Sun, Z-M., Fu, G., Galanin, DA., Nagasato, C., Motomura, T., Hu, Z-M. & Duan, D-L. 2015. Phylogeographic data revealed shallow genetic structure in the kelp Saccharina japonica (Laminariales, Phaeophyta). BMC Evol Biol. 15:237 pp.
|
|
|||||||||||||||||||||||||||||||||||||||||