INTRODUCTION
The genus Umbraulva was segregated from the genus Ulva by Bae and Lee (2001) and includes three species previously described as U. amaniensis Tanaka (type locality: Amami-Oshima, Japan), U. japonica (Holmes) Papenfuss (type locality: Enoshima, Japan), and U. olivascens Dangeard (type locality: Roscoff, France). Umbraulva is distinguished from Ulva Linnaeus by its distinct olive-green blade, subtidal habitat, and the presence of the pigment siphonaxanthin (Bae and Lee 2001), which pigment can grow in deeper waters because they absorb green light in the 540-nm range (Yokohama 1981). Umbraulva kuaweuweu H. L. Spalding & A. R. Sherwood and Um. kaloakulau H. L. Spalding & A. R. Sherwood were described as new species from specimens collected at depths of 80–125 m in mesophotic ecosystems of the Hawaiian Archipelago based on vegetative morphology and molecular phylogenetic analyses of the internal transcribed spacer (ITS) region, ribulose-1,5-biphosphate carboxylase large subunit (rbcL), and elongation factor Tu (tufA) genes (Spalding et al. 2016). Although it was known that Umbraulva includes five species taxonomically (Guiry and Guiry 2020), the new genus Ryuguphycus H. Kawai, T. Hanyuda & T. Kitayama, which includes R. kuaweuweu (formerly Um. kuaweuweu) as the generitype, was recently separated from the genus Umbraulva based on its distinctive morphology, life history, carotenoid composition, and molecular phylogeny (Kawai et al. 2020).
Two Umbraulva species, Um. amamiensis and Um. japonica, have been reported from subtidal habitats in Korea (National Institute of Biological Resources 2019); however, it is very difficult to identify species correctly based on a few traditional morpho-anatomical characteristics such as blade shape and thickness, presence / absence of microscopic marginal denticulation, and rhizoidal filament type (Spalding et al. 2016, Kawai et al. 2020). The plastid markers rbcL and tufA have been used to delimit species boundaries in Umbraulva, including several new undescribed specimens (Heesch et al. 2009, Kirkendale et al. 2013). Therefore, it is necessary to study the systematics of this genus using molecular phylogenetic information and/or monitor the occurrence of invasive species in subtidal areas. During subtidal zone surveys around Jeju Island, Korea, we discovered a novel Umbraulva species; this finding is supported by molecular analyses and morphological observations. The objectives of this study were to determine the taxonomic position of this new species, assess its genetic differences from related taxa, and discuss the phylogenetic relationships of Umbraulva based on the plastid rbcL and tufA genes.
MATERIALS AND METHODS
Using scuba gear, we collected six specimens of a new Umbraulva candidate and four of Um. japonica from depths of 8–15 m in subtidal areas around four annexed islets (Hyeongjeseom, Munseom, Seopseom, and Udo) of Jeju Island, Korea. The fresh samples were photographed using an Olympus TG-4 waterproof digital camera (Olympus, Tokyo, Japan). Thallus fragments of each specimen were dried for molecular analyses using silica gel. Prior to describing morphological characters, the samples were preserved in 5% formalin in seawater and sectioned using a bench-top freezing microtome (NK-101-II; Nippon Optical Works Co., Ltd., Tokyo, Japan). Sectioned materials were stained with 1% aniline blue acidified with 1% HCl after bleaching under sunlight. Sections were mounted in 35% corn syrup and photographed under a microscope (BX43; Olympus) using an EOS 600D digital camera (Canon, Tokyo, Japan). Digitized images were edited for clarity using Adobe Photoshop software (ver. 6.1; Adobe Systems Inc., San Jose, CA, USA). Pressed herbarium specimens were deposited as voucher specimens in the herbaria of Jeju National University (JNUB) and the National Institute of Biological Resources (KB), Incheon, Korea.
Total genomic DNA of two Umbraulva species (Table 1) was extracted following the protocol of the LaboPass Tissue Genomic DNA Isolation Kit Mini (Cosmo Genetech, Seoul, Korea). All polymerase chain reaction (PCR) processes were performed using AccuPower PCR Premix (Bioneer, Daejeon, Korea), following the manufacturer’s protocol. We amplified and sequenced the plastid genes rbcL and tufA and analyzed nuclear 18S rRNA from an unidentified Umbraulva species. In the present study, the primer combinations for rbcL, tufA, and 18S rRNA-5P were GrbcLFi (Saunders and Kucera 2010) or RH1 (Manhart 1994) / 1385R (Manhart 1994), tufGF4 / tufGR (Saunders and Kucera 2010), and SSU-A (Medlin et al. 1988) / SSU_inR (Manhart 1994), respectively. The PCR amplification procedure followed that of Saunders and Kucera (2010). All successfully amplified PCR products were purified using an AccuPrep PCR Purification Kit (Bioneer) and sequenced by Macrogen (Seoul, Korea) using forward and reverse primers. Additional rbcL and tufA sequences for the phylogenetic analysis were selected from GenBank (Supplementary Table S1). The rbcL and tufA datasets were aligned visually using the BioEdit software (Hall 1999) after editing the Umbraulva sequences obtained in this study using Chromas ver. 1.45 software (Technelysium Pty Ltd., South Brisbane, Australia). Variations in the rbcL and tufA sequences were assessed based on uncorrected pairwise genetic distances (p-distance) using MEGA 5.1 software (Tamura et al. 2011) and a neighbor-joining algorithm dependent on the Kimura two-parameter distance method. To determine the taxonomic positions of our Umbraulva specimens (Table 1), each rbcL and tufA phylogeny was run using a maximum-likelihood (ML) algorithm with RAxML software (Stamatakis 2006) and the GTR + Γ + I model with 1,000 bootstrap (BS) replicates from each sequence dataset for our Umbraulva specimens (Table 1) and related ulvacean taxa (Supplementary Table S1), including Ulvaria obscura (rbcL, HQ603436 and HQ603651; tufA, HQ610415) as an outgroup. We constructed a combined phylogenetic tree inferred from the rbcL, tufA, 18S rRNA, 28S rRNA, and ITS sequence datasets (Supplementary Table S1) to delimit the generic boundaries within Ulvaceae. This concatenated phylogenetic tree was constructed using the ML method in RAxML software and expressed using FigTree ver. 1.4.0 software (Rambaut 2012).
RESULTS
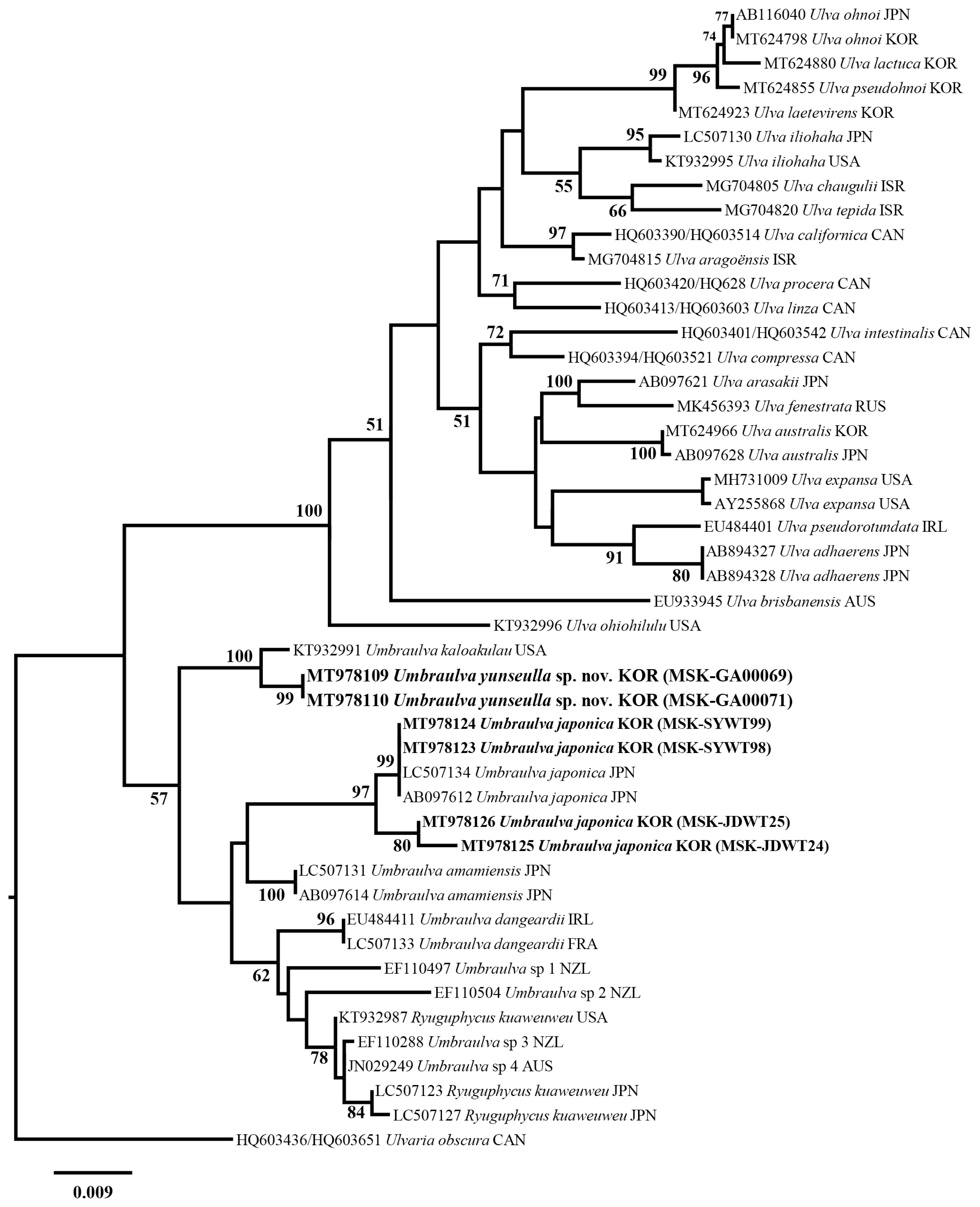
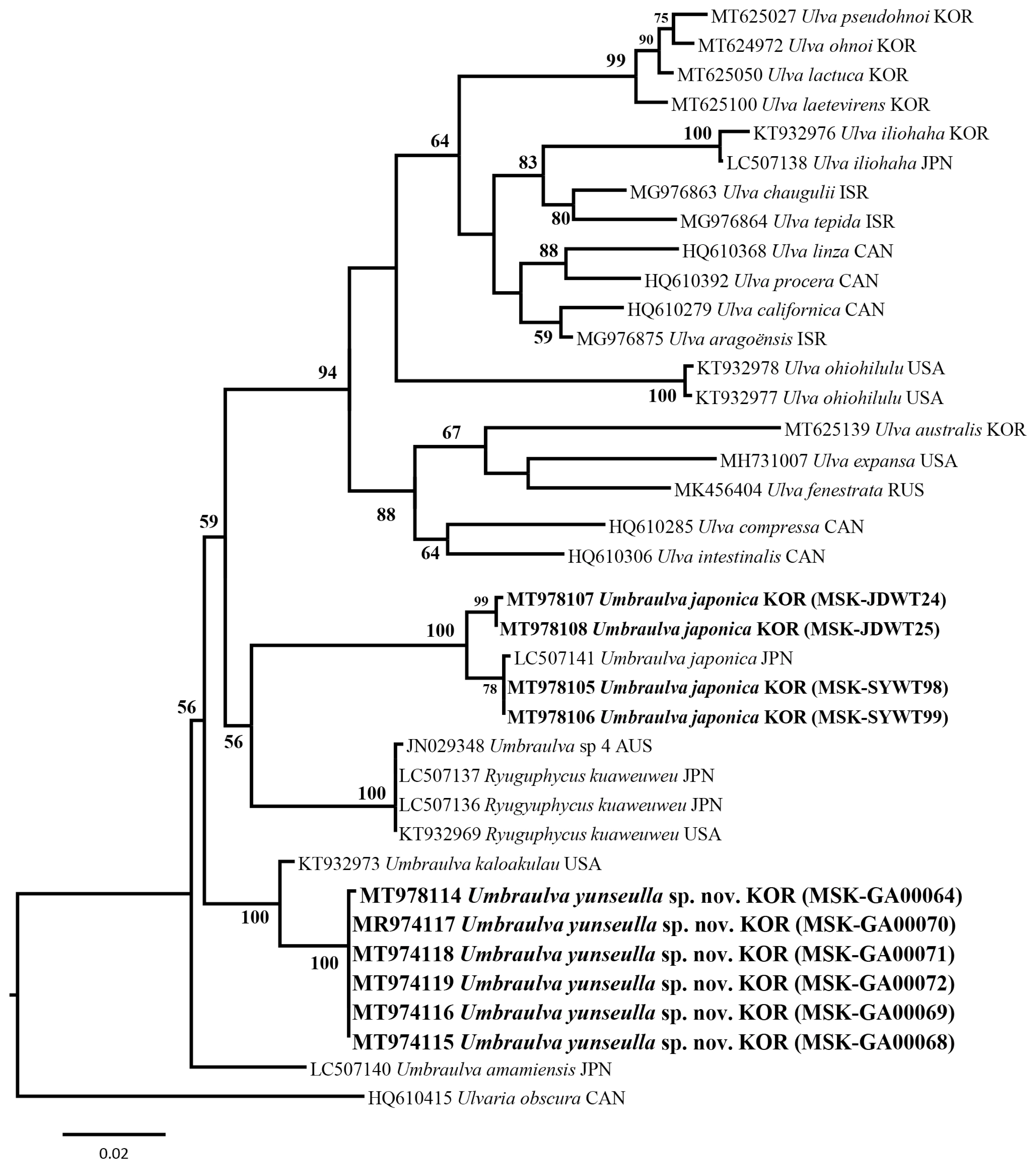
We analyzed a set of 53 rbcL gene sequences including two new Umbraulva sp. specimens (MT978109–MT978110; 741–745 bp) and four Um. japonica ones (MT978123–MT978126; 1,263–1,273 bp). Among all sites rbcL, 185 sites (14.5%) were variable and 137 sites (10.8%) were parsimoniously informative. In tufA analysis, a set of 40 tufA gene sequences were analyzed including six new Umbraulva sp. specimens (MT978114–MT978119; 804–859 bp) and four Um. japonica ones (MT978107–MT978106; 848–866 bp). Variable and parsimoniously informative sites were 327 sites (37.8%) and 169 (19.7%), respectively. In addition, a 18S rRNA-5P sequence were obtained from a new Umbraulva sp. specimen (MT978101; 530 bp). Phylogenetic analysis of the plastid rbcL and tufA sequences showed that the six new Umbraulva sp. specimens from Jeju Island were separated from other species within Umbraulva clade in a fully supported clade (BS, 100), with sister to Um. kaloakulau from Hawaii, USA (Figs 1 & 2). The rbcL phylogeny showed that the novel Umbraulva sp. had diverged from the Um. japonica clade from Korea (MT978123, MT978124, MT978125, and MT978126) including the generitype from Japan (AB097612 and LC507134), as well as Um. amamiensis (AB097614 and LC 507131), with an interspecific variation of 2.6–3.3% and 2.6%, respectively (Fig. 1). Higher interspecific divergence among Umbraulva species was observed in the tufA phylogenentic analysis compared to the rbcL results. The tufA interspecific variation values for the novel Umbraulva sp. were 1.6% vs. Um. kaloakulau, 6.6–7.0% vs. Um. japonica, and 4.2% vs. Um. amamiensis.
Although the genus Ulva is monophyletic based on the rbcL and tufA genes ML phylogeny, Umbraulva clade was not monophyletic because of the genus Ryuguphycus embedded within this clade. In addition, the concatenated ML phylogeny showed that Umbraulva consists of two clades, one containing Um. japonica, Um. amamiensis, Um. kaloakulau, and the novel Umbraulva sp. (Fig. 3) with weak ML support (BS, 69%), and the other consisting of Um. dangeardii from Ireland and France (EU484411 and LC507133, respectively), Umbraulva sp. specimens from New Zealand (EF110497, EF110504, and EF110288) and Australia (JN029249, rbcL, and JN029348) (Supplementary Table S1), and Ryuguphycus with no support (Fig. 3). Although the clade of Umbraulva is unstable on the basis of the molecular evidence, we propose a new species of Umbraulva, hereby named Um. yunseulla H. W. Lee, E. H. Bae & M. S. Kim sp. nov.
Umbraulva yunseulla H. W. Lee, E. H. Bae & M. S. Kim sp. nov. (Fig. 4A–L)
Holotype
MSK-GA00069 (Fig. 4B), vegetative, Munseom (annexed islet), Jeju Island, Korea, Jan 30, 2013, deposited in the JNUB herbarium: GenBank accession No. MT978109, rbcL; MT978116, tufA.
Isotypes
MSK-GA00068, MSK-GA00071, vegetative, Munseom (annexed islet), Jeju Island, Korea, Jan 30, 2013 (deposited in JNUB). MSK-GA00064, MSK-GA00070 (Fig. 4C), MSK-GA00072, vegetative, Munseom, Jeju Island, Korea, Jan 30, 2013 (deposited in KB).
Etymology
The specific epithet, yunseulla, is derived from the Korean traditional noun yunseul, meaning “calm wave glittering with sunshine or moonlight” in reference to the greenish iridescence observed on the underwater thallus surface.
Habitat
Attached to hard substrates such as rocks, shells, or nongeniculate corallines in the subtidal zone (generally 8–15 m deep).
Specimens examined
MSK-GA00064, MSK-GA00068, MSK-GA00069, MSK-GA00070, MSK-GA00071, MSK-GA00072, Jan 30, 2013, Munseom, Jeju Island, Korea; MSK150802-06, Aug 2, 2015, Sagye, Jeju Island, Korea; MSKL160414-14, MSKL160414-15, MSKL160414-16, MS-KL160414-17, Apr 14, 2016, Munseom, Jeju Island, Korea; MSKL160419-03, MSKL160419-12, MSKL160419-13, MSKL160419-14, Apr 16, 2016, Munseom, Jeju Island, Korea; MSKL160419-19, MSKL160419-22, Apr 19, 2016, Seopseom (annexed islet), Jeju Island, Korea; MSKL160520-10, May 20, 2016, Munseom, Jeju Island, Korea; MSKL160528-16, MSK160528-17, May 28, 2016, U-do (annexed islet), Jeju Island, Korea (deposited in JNUB).
DNA sequence data
rbcL: MSK-GA00069 (MT978109), MSK-GA00071 (MT978110). tufA: MSK-GA00064 (MT978114), MSK-GA00068 (MT978115), MSK-GA00069 (MT978116), MSK-GA00070 (MT978117), MSK-GA00071 (MT978118), MSK-GA00072 (MT978119). 18S rRNA: MSK-GA00064 (MT- 978101).
Habit and morphology
Umbraulva yunseulla sp. nov. has a foliose (Fig. 4B & C) and distromatic (Fig. 4H & I) thallus with slightly ruffled to curled margins (Fig. 4B & C). Greenish iridescence derived from the natural habitat glitters on the surface radially from the lower part or at the margin of the thallus (Fig. 4A). Thallus green to dark green (Fig. 4B & C). Thallus entirely globular to subglobular and funnel-shaped, growing dorsiventrally decumbent when young to erect when mature (Fig. 4A), 5–7 cm wide and 4–5 cm high, to a maximum of 10 cm (Fig. 4B & C). Thallus apex rounded to slightly emarginated, base cordate to lobate (Fig. 4B & C). Thallus attached by a distinct discoid holdfast without a stipe (Fig. 4B & C) and generally composed of a single or 2–3 (5) blades (Fig. 4B & C). Distromatic thallus is composed of entirely cuboidal to polygonal cells, some subspherical in the upper part of the blade in surface view, 11–26 μm long by 9–16 μm wide (Fig. 4D). In the center of the blade, cells are subspherical, cuboidal to polygonal, 15–35 μm long by 9–18 μm wide, and are larger and more numerous than in the upper blade (Fig. 4E). At the base, subspherical, cuboidal and polygonal cells, 18–55 μm long by 9–26 μm wide, are intermixed (Fig. 4F). Pale cells are greatly expanded, and darker cells are filled with cytoplasm in a compact arrangement (Fig. 4F). Each cell contains 1–2 pyrenoids (Fig. 4E), up to 4 per cell. Cell arrangement throughout the thallus irregular (Fig. 4D–F). Thallus margin entire, plain, and rounded, without microscopic protuberances (Fig. 4G). In transverse section, cell shape is cuboidal throughout the entire blade (Fig. 4H–J). Cell size in transverse section is taller and wider toward basal part, 26–32 μm tall by 11–13 μm wide in upper, 23–28 μm tall by 13–17 μm wide in middle, 40–60 μm tall by 23–37 μm wide in basal, but similar throughout the thallus on both the dorsal and ventral surfaces (Fig. 4H–J). Cell thickness ranges from 50–70 μm in the upper thallus to 100–140 μm in the basal portion of the blade. In transverse section, the basal portion of the blade, fine rhizoidal filaments are observed budding off from cells inward filled with cytoplasmic contents compactly, and are interwoven with longitudinal arrangement and fill between distromatic cell planes densely (Fig. 4J–L). Gametophytes were not observed.
DISCUSSION
Molecular analyses of the genus Umbraulva from subtidal areas of Jeju Island, Korea, have expanded our understanding of the phylogenetic affinity between Umbraulva and related taxa (Bae and Lee 2001). This approach has facilitated the discovery of the newly recognized species Um. yunseulla sp. nov., thus enhancing studies of macroalgal biodiversity. The plastid markers rbcL and tufA were previously demonstrated to resolve new species and delimit boundaries among Umbraulva species (Heesch et al. 2009, Kirkendale et al. 2013). In this study, the morphological delimitation, such as size and thickness of thallus, size and shape of cell, and number of pyrenoids, provide less resolution to identify species definitely because morphological characteristic range is overlapped between Umbraulva species (Table 2). However, the phylogenetic analyses inferred rbcL and tufA are helpful to reveal a new member of Umbraulva, Um. yunseulla sp. nov. from Jeju Island, Korea (Figs 1 & 2), which has the smallest and a globular to sub-gulobular and funnel-shaped thallus with greenish iridescence along dorsal surface (Table 2).
Umbraulva yunseulla sp. nov. was first collected by Bae and Lee (2001) from Munseom, Jeju Island, Korea, but it was identified as Um. amamiensis, which is described as having a height of 20–60 cm (to 115 cm) and width of 10–35 cm (to 55 cm), with abundant perforations (Tanaka 1956). Um. yunseulla sp. nov. has much smaller thallus, ranged 5–7 cm wide and 4–5 cm high up to a maximum of 10 cm, compared to Um. amamiensis, although these two species inhabit the similar depth range of subtidal (Table 2). The specimens of Bae and Lee (2001) were characterized by cordate or funnel-shaped thalli 5–7 cm in height with wavy margins and epifluorescent illumination at the surface of the blade in nature. These characteristics are identical to our specimens of Um. yunseulla sp. nov. (Table 2). On the contrary, Um. amamiensis is linear lanceolate to ovate-lanceolate with irregular perforation, and the epifluorescent illumination of Um. amamiensis specimens was undescribed (Table 2). The iridescent coloration along thallus surface is one of morphological features of Um. yunseulla sp. nov. distinguished from other Umbraulva species (Table 2). In marine macroalgae, external iridescence is caused by unique nanostructural coloration mechanisms such as intracellular iridescent bodies or multi-layered cuticle reflection (Chandler et al. 2017). Some brown and red seaweeds such as Cystoseria, Dictyota, Chondria, and Cottoniella have nanostructural coloration that produces iridescent bodies (Chandler et al. 2017). In Um. yunseulla sp. nov., greenish iridescence appears around the base or middle part of the thallus (Fig. 4A). In surface view, Um. yunseulla sp. nov. exhibits larger subspherical and expanded cells than it does cuboidal or polygonal cells; these are mainly distributed at the middle and basal parts of the thallus, and have tiny globules scattered throughout (Fig. 4E & F). Further investigation may show that these tiny globules are iridescent bodies of Um. yunseulla sp. nov., which would provide new insight into the mechanism by which iridescent marine macroalgal species adapt to environmental conditions such as radiation intensity and turbidity (Chandler et al. 2017).
The monophyly of Umbraulva was not well-supported by rbcL and tufA phylogenetic analyses (Figs 1 & 2), which is inconsistent with a previous study of the phylogenetic relationship between Umbraulva and Ryuguphycus (Kawai et al. 2020). Because the generic boundary of Umbraulva was delimited by only weak support and the Ryuguphycus clade was supported by poor BS support in our concatenated analyses inferred from 18S rRNA, 28S rRNA, ITS, rbcL, and tufA (Fig. 3), the phylogenetic topology between Umbraulva and Ryuguphycus presented in this study remains uncertain. The recently established genera Umbraulva and Ryuguphycus have been subject to fewer phylogenetic analyses than Ulva, which has been examined using many molecular approaches (Kirkendale et al. 2013, Spalding et al. 2016). Future studies should attempt to discover new members of Umbraulva and Ryuguphycus to fill phylogenetic gaps and strengthen generic boundaries among these genera.
Few taxonomic studies have examined ulvacean organisms inhabiting subtidal zones of Korea, with the exception of Umbraulva species (Bae and Lee 2001). Recent studies have delimited species boundaries within the genus Codium Stackhouse, including five subtidal species, and for Palmophyllum crassum (Naccari) Rabenhorst, which inhabits deep marine areas around Jeju (Lee and Kim 2015, 2017). Many red algae species have been discovered in the subtidal zones of Jeju Island, including Pseudopolyneura hyacinthia (J. C. Kang & M. S. Kim) M. J. Wynne (as Erythroglossum hyacinthinum, Kang and Kim 2014) and Pachymeniopsis volvita M. Y. Yang & M. S. Kim (Yang and Kim 2015). Although Jeju Island has the potential to be named a seaweed genetic and species diversity hotspot (Yang et al. 2020), our knowledge of green seaweeds in subtidal zones remains poor. A few new ulvacean species have been reported in Hawaii and Japan, including U. oiohilulu, Um. kaloakulau, and R. kuaweuweu collected from the mesophotic zone, at depths of 30–125 m (Spalding et al. 2016, Kawai et al. 2020). Continuous surveys of green algae in the subtidal zone contribute to the expansion of ecological knowledge, which is essential for habitat conservation and species diversity enrichment. Therefore, we further emphasize the necessity of investigating green macroalgal diversity in the mesophotic zone to discover new species of the ulvaceaen genera Ulva, Umbraulva, and Ryuguphycus.