ABSTRACT
Haraldiophyllum hawaiiense sp. nov. is described as a new mesophotic alga and a new genus record for the Hawaiian Islands. Six specimens were collected at a depth range of 81–93 m from Papahānaumokuākea Marine National Monument, and their morphology investigated, as well as molecular phylogenetic analyses of the plastidial ribulose-1,5-bisphosphate carboxylase–oxygenase large-subunit (rbcL) gene and a concatenated alignment of rbcL and nuclear large-subunit rRNA gene (LSU) sequences. Phylogenetic analyses supported H. hawaiiense sp. nov. as a distinct lineage within the genus Haraldiophyllum, and sister to a large clade containing the type species, H. bonnemaisonii, as well as H. crispatum and an undescribed European specimen. The six Hawaiian specimens were shown to be identical, but unique among other species of the genus as well as the recently segregated genus Neoharaldiophyllum, which comprises half of the species previously included in Haraldiophyllum. The vegetative morphology of H. hawaiiense sp. nov. resembles Neoharaldiophyllum udoense (formerly H. udoensis); however, no female or post-fertilization structures were found in the Hawaiian specimens to allow a more comprehensive comparison. The molecular phylogenies demonstrate that Haraldiophyllum is paraphyletic, suggesting either that the Myriogrammeae tribe includes undescribed genera, including Haraldiophyllum sensu stricto, or that Neoharaldiophyllum species should be transferred into the genus Haraldiophyllum. However, based on vegetative morphology and molecular analyses, and pending resolution of this taxonomic issue, the Hawaiian specimens are placed within the genus Haraldiophyllum. This new record for the Hawaiian Islands highlights the novel biodiversity from mesophotic depths, reaffirming the need for further investigation into the biodiversity of Mesophotic Coral Ecosystems.
INTRODUCTIONThe delesseriacean genus Haraldiophyllum was established by Zinova (1981) to include three previously described species: Nitophyllum bonnemaisonii (Kylin) Kylin (= Myriogramme bonnemaisonii Kylin) and N. versicolor Harvey from Atlantic Europe, and N. mirabile Kylin from Washington, USA. In these three species the two pericentral cells that are cut off from the fertile central cell in the procarp are located one on top of the other (i.e., parallel to the longitudinal axis of the thallus), while in the generitype of Nitophyllum, N. punctatum (Stackhouse) Greville (Kylin 1934), the pericentral cells are organized perpendicular to the axis (Wynne 2014, Kang et al. 2017). Furthermore, N. punctatum is characterized by a single sterile-cell group in the development of the procarp, while the other three species have two sterile-cell groups (Kylin 1925, 1934). Four other species have been assigned to Haraldiophyllum, yielding seven species in total: H. bonnemaisonii (Kylin) A. D. Zinova (designated the generitype), H. crispatum (J. D. Hooker & Harvey) Showe M. Lin, Hommersand & W. A. Nelson, H. erosum (Harvey) A. J. K. Millar & Huisman, H. infossum A. J. K. Millar, H. mirabile (Kylin) A. D. Zinova, H. nottii (R. E. Norris & M. J. Wynne) M. J. Wynne, and H. sinuosum (A. H. S. Lucas) A. J. K. Millar. Haraldiophyllum udoensis M. S. Kim & J. C. Kang, which was previously misidentified in Korea as H. bonnemaisonii (Nam and Kim 1996), was described as a new species based on vegetative morphological comparisons and phylogenetic analysis of the ribulose-1,5-bisphosphate carboxylase-oxygenase large-subunit (rbcL) gene (Kim and Kang 2011). In 2017, Kang and Kim proposed the genus Neoharaldiophyllum, establishing N. udoense (M. S. Kim & J. C. Kang) J. C. Kang & M. S. Kim as the generitype based on phylogenetic analyses of rbcL and nuclear large-subunit rRNA gene (LSU) markers and the following combination of morphological features: the primary gonimoblast filaments are secondarily fused with many floor cells of the cystocarp cavity, and arrangement of tetrasporangia within the tetrasporangial sori is symmetrical in two layers (Kang et al. 2017). They also transferred three other species of Haraldiophyllum to Neoharaldiophyllum, such that the genus currently contains N. erosum (Harvey) J. C. Kang & M. S. Kim, N. mirable (Kylin) J. C. Kang & M. S. Kim, N. nottii (R. E. Norris & M. J. Wynne) J. C. Kang & M. S. Kim and N. udoense.
Haraldiophyllum and Neoharaldiophyllum species have been recorded worldwide, with H. bonnemaisonii reportedly having the widest distribution, including several locations in Europe, as well as the Atlantic Islands and the Middle East (Guiry and Guiry 2020). The remaining species of both genera have more limited distributions, for example H. infossum, which occurs only along the eastern coast of Australia and New Zealand (Guiry and Guiry 2020). There are no records of either genus for the Hawaiian Archipelago to date.
Over the past 15 years, thousands of algal specimens have been collected during expeditions to Pa-pahānaumokuākea Marine National Monument (PMNM) led by the US National Oceanic and Atmospheric Administration (NOAA), as part of an ongoing effort to characterize the algal biodiversity of Hawaiian Mesophotic Coral Ecosystems (MCEs). Here we describe a new species, Haraldiophyllum hawaiiense sp. nov., which is also a new genus record for the Hawaiian Islands, based on morphological and molecular characterization of six specimens collected in 2015 and 2019 from PMNM.
MATERIALS AND METHODSSix Haraldiophyllum specimens were collected during technical diving operations on cruises to PMNM between 2015 and 2019 (Table 1). Four specimens were collected from Kure Atoll (Hōlanikū), and one specimen each was collected from Pearl & Hermes Atoll (Manawai), and Salmon Bank, at a depth range of 82 to 93 m. Specimens were preserved in 4% formalin for morphological studies and in silica gel desiccant for molecular analyses. Voucher specimens were mounted on herbarium paper and accessioned at the Bernice P. Bishop Museum (BISH) under the accessions BISH 776069–776071 and BISH 776120–776122. Morphological and anatomical investigations of the samples were conducted by gently rehydrating the thalli in a detergent (Modified Pohl’s Solution, Clark, unpublished: https://www.eeob.iastate.edu/research/bamboo/pdf/anatomy_protocols.pdf) for at least 2 h, hand sectioning with a double-edged razor blade, staining with 0.5% aniline blue (Figs 6 & 13 were not stained), and mounting in 30% Karo (ACH Food Companies, Inc., Memphis, TN, USA). Photomicrographs were taken on a Zeiss AxioImager A1 compound light microscope (Pleasanton, CA, USA) with an Infinity2-1RC digital camera (Lumenera Corporation, Ottawa, ON, Canada).
Genomic DNA was extracted using the OMEGA E.Z.N.A. Plant DNA DS Kit (OMEGA Biotek, Norcross, GA, USA) protocol. The genes selected for the molecular analyses were the chloroplast-encoded rbcL and the nuclear-encoded LSU. The rbcL gene was amplified using primers for two overlapping fragments: rbcLF7 and rbcLJNR1 (Gavio and Fredericq 2002, Kang and Kim 2013) for the first fragment, and rbcLF762 and rbcLR1442 (Kim et al. 2010) for the second fragment. A 1,390-bp fragment of LSU was amplified using the following overlapping primer pairs: T16 and T24, and T25 and T10 (Harper and Saunders 2001). A 630-bp fragment from the DNA barcode region near the 5′ end of the mitochondrial cytochrome oxidase subunit I (COI) gene was generated using the GazF1 and GazR1 primer pair (Saunders 2005). Polymerase chain reaction products that amplified successfully were sent to be sequenced at the University of Hawai‘i at Mānoa Advanced Studies in Genomics, Proteomics, and Bioinformatics (ASGPB) core facility. To date, there are no other COI sequences for Haraldiophyllum on GenBank and only a single sequence on Barcode of Life Database (BOLD) (ABMMC218554-16, H. bonnemaisonii); the COI sequence generated in the current study was aligned to this sequence from BOLD for comparison. For phylogenetic analyses of the rbcL and LSU sequences, raw sequences were assembled and aligned using the MUSCLE v. 3.8.425 plug-in (Edgar 2004) in Geneious Prime 2019.1.3 (http://www.geneious.com), and edited manually where necessary. Two alignments were generated: one for rbcL and a concatenated alignment for rbcL + LSU. For each, contiguous sequences were aligned with reference sequences and analyzed with PartitionFinder v. 1.1.1 (Lanfear et al. 2012). Maximum likelihood (ML) analysis was conducted on both alignments using RAxML-HPC2 on XSEDE v. 8.2.10 (Stamatakis 2014) via the CIPRES gateway (Miller et al. 2010) with 1,000 bootstrap replicates and the GTRCAT model. Bayesian inference analysis (BA) was performed using the MrBayes plug-in v. 3.2.6 (Huelsenbeck and Ronquist 2001) through Geneious Prime 2019.1.3 (http://www.geneious.com) using four chains of Metropolis-coupled Markov Chain Monte Carlo for 1,000,000 generations and with sampling every 100 generations; 100,000 chains were removed as burn-in prior to determining posterior probabilities.
RESULTSComparison of the COI barcode sequences from the Hawaiian specimens ARS 09801, ARS 09803, and ARS 09938 to those in GenBank and BOLD returned the following values: 91.0% and 90.7% similarity with H. bonnemaisonii (BOLD: ABMMC21854-16) from Norway and N. mirable from Canada (BOLD: ABMMC1053-06), respectively, and 90.8% similarity with eight Neoharaldiophyllum udoense specimens (GenBank: KY497914–KY497921) from Korea (Kang et al. 2017). Haraldiophyllum currently includes four species, but only the type, H. bonnemaisonii, has a COI barcode sequence; for this reason, a neighbor-joining analysis was not performed.
Phylogenetic analyses were conducted using the rbcL and rbcL + LSU concatenated alignments of Haraldiophyllum and closely related taxa belonging to the subfamily Phycodryoideae. Two genera of the subfamily Nitophylloideae were included as outgroups: Augophyllum delicatum (A. J. K. Millar) Showe M. Lin, Fredericq & Hommersand and Nitophyllum punctatum (Stackhouse) Greville, which were obtained from GenBank (Supplementary Table S1). The rbcL alignment was 1,442 bp in length, of which 547 sites (37.8%) were parsimony informative. The concatenation of LSU + rbcL sequences yielded an alignment of 2,813 bp, with 1,156 sites (39.6%) parsimony informative. The rbcL sequence divergences between species of Haraldiophyllum and Neoharaldiophyllum ranged from 2.5–7.0%, whereas the Hawaiian sequences had divergences with H. crispatum and Haraldiophyllum sp. 2 of 5.5% and 7.0%, respectively. Comparing the Hawaiian rbcL sequences with those of Neoharaldiophyllum demonstrated a slightly lower divergence with N. udoense (5.2%) and N. mirable (6.3%). In the concatenated phylogeny, the Hawaiian sequences diverged by 5.4–6.1% from H. crispatum and H. bonnemaisonii, respectively, and by 5.3–6.7% from Haraldiophyllum sp. from Chile and N. mirable, respectively. ML and BA analyses for both alignments yielded similar tree topologies, and so only the ML results are shown, with support values from both ML bootstrap and BA posterior probabilities displayed. In the rbcL phylogenetic tree (Fig. 1), the tribes Myriogrammeae and Phycodryeae were clearly separated, and the new Haraldiophyllum species was sister to a clade with H. bonnemaisonii, H. crispatum, and “Haraldiophyllum sp. 1.” This Haraldiophyllum clade was sister to Neoharaldiophyllum, and a third clade, consisting of four sequences of “Haraldiophyllum sp. 2”, was sister to this combined Haraldiophyllum + Neoharaldiophyllum clade; thus, the genus Haraldiophyllum is not resolved as monophyletic in these analyses. In the combined LSU and rbcL tree (Fig. 2), the Haraldiophyllum clade included the new Hawaiian Haraldiophyllum species, as well as the type species H. bonnemaisonii and H. crispatum, with good support (80% bootstrap and 0.98 posterior probability), again forming a sister clade to Neoharaldiophyllum. The divergence levels demonstrated that the Hawaiian specimens likely represent a distinct species, which is described below. Note that Haraldiophyllum sp. from Chile is described as “Haraldiophyllum ramirezii” on GenBank, under accession numbers AF254186–AF254188 for rbcL, and AF259430 for LSU. In all references (including the authors’ publication), these sequences cluster with Neoharaldiophyllum, and are mentioned only as Haraldiophyllum sp.; furthermore, there is no mention of the species name “Haraldiophyllum ramirezii” on AlgaeBase or in BOLD.
Haraldiophyllum hawaiiense M. O. Paiano, Huisman & A. R. Sherwood sp. nov. (Fig. 3)DescriptionPlants erect, epilithic, or as epiphytes on other algal species. Thalli irregularly or dichotomously branched, composed of one to numerous ovate to elongate blades, attached by a small rounded holdfast, with occasional polystromatic basal nerves, 425–559 μm thick, with 8–10 cell layers. Thalli rose-pink to brownish-red, 2.2–7.8 cm high and 2.2–13.2 cm broad. Blades with a delicate texture, denticulate margins, monostromatic at the tips and margins. Blades in surface view composed of polygonal cells, 50–94 μm long × 75–106 μm wide, presenting parietal granular chloroplasts; cells are connected by primary and secondary pit connections. Vegetative growth by transverse division of marginal cells. Tetrasporangial sori circular to oval, 300–488 μm long by 238–475 μm wide, scattered throughout both sides of the blade, except along the margins, up to 210 μm thick. Mature tetrasporangia spherical to slightly ovate, 43–67 μm long by 45–68 μm in diameter, tetrahedrally divided. Gametophytes not observed.
HolotypeBISH 776069 (ARS 09803, Kure Atoll (Hōlanikū), Hawai‘i, 28.49007, −178.29113, 88 m depth, Sep 21, 2015, NWHI 455, coll. R. Kosaki & B. Hauk). GenBank accession Nos. MT161613 (rbcL), MT161615 (LSU), MT161617 (COI).
TopotypeBISH 776122 (ARS 09985, Kure Atoll (Hōlanikū), Hawai‘i, 28.44342, −178.26172, 87 m depth, Sep 21, 2015, NWHI 443a, coll. D. Wagner).
DistributionMesophotic depths at Kure Atoll (Hōlanikū) (88–91 m depth), Salmon Bank (82 m depth), and Pearl & Hermes Atoll (Manawai) (93 m depth), Hawai‘i, USA.
Specimens examinedBISH 776069 (ARS 09803), BISH 776070 (ARS 09802), BISH 776071 (ARS09801), BISH 776120 (ARS 09938), BISH 776121 (ARS 09984), and BISH 776122 (ARS 09985).
DNA sequence dataGenBank accession Nos. MT-161616–MT161618 (COI), MT161614–MT161615 (LSU), and MT161612–MT161613 (rbcL).
Habit and vegetative morphologyAll specimens were collected from the mesophotic coral ecosystem (81–93 m) in the PMNM. Plants are erect, up to 7.8 cm high and 13.2 cm in breadth, epilithic, attached by a small rounded holdfast or as epiphytes on other algal species. As examples, ARS 09984 was found associated with the holdfast of a brown alga, Sporochnus, while ARS 09985 was an epiphyte on an unknown red alga, attached by creeping stolons. When alive, plants were rose-pink, and turned to brownish-red when dry. Thalli are composed of one to many membranous blades (Fig. 3A & B), arising from the holdfast, or creeping stolons when epiphytic, blades frequently cleft with denticulate margins (Fig. 3C), with an obtuse or apiculate apex (Fig. 3D). The blades are thin and mostly monostromatic (Fig. 3E), with no midribs or microscopic veins on the blades except for the lower portion of some blades where occasionally a fan-shaped thickened nerve was observed, becoming polystromatic (Fig. 3F & G). Marginal growth occurs by transverse or oblique divisions. Tetrasporangial sori are circular to elliptical (Fig. 3D & H), numerous and randomly scattered on both sides of the blade surface (Fig. 3D), close to the margins and upper parts of the blade. Tetrasporangial sori protrude on either side of the blade and are produced from inner and subcortical cells; in transverse section tetrasporangia are clearly visible in two layers between both surface cells, separated by a central medullary cell layer (Fig. 3J). Mature tetrasporangia are spherical, 43–68 μm in diameter and divided tetrahedrally (Fig. 3I), with numerous tetrasporangia formed within a single sorus (Fig. 3K). No male or female reproductive structures were observed.
DISCUSSION
Haraldiophyllum (Zinova 1981), includes H. bonnemaisonii (Kylin) A. D. Zinova [= Myriogramme bonnemaisonii Kylin], as the type species, H. crispatum (J. D. Hooker & Harvey) S. -M. Lin, Hommersand & W. A. Nelson, H. infossum A. J. K. Millar, and H. sinuosum (A. H. S. Lucas) A. J. K. Millar, the last two being previously assigned to Myriogramme and Nitophyllum, respectively. The genus was erected as one of several taxonomic changes in the family Delesseriaceae proposed to solve a misunderstanding created by Kylin (1924) when he described Myriogramme based on two different taxa: M. livida (J. D. Hooker & Harvey) Kylin and M. minuta Kylin. The genus Neoharaldiophyllum was raised by Kang et al. in 2017 with N. udoense as the type, and was differentiated from Haraldiophyllum by the primary gonimoblast being secondarily fused with floor cells of the cystocarp (versus being free, without secondary fusions in Haraldiophyllum) and a symmetrical arrangement of tetrasporangia in two layers (as opposed to the asymmetrical presentation in Haraldiophyllum). The number of layers in the tetrasporangial arrangement does not seen to be a significant character for genera within the Myriogrammeae, varying as one or two layers within Haraldiophyllum and Neoharaldiophyllum, versus two layers in Myriogramme and Platyclinia (Maggs and Hommersand 1993, Lin et al. 2007, Kim and Kang 2011, Kang et al. 2017).
Within the tribe Myriogrammeae, which was described by Hommersand and Fredericq (1997), Haraldiophyllum, Neoharaldiophyllum and Myriogramme are very similar in morphology, and the following features characterize the tribe: procarps associated with anterior cover cells, and consisting of a supporting cell, a one- to two-celled lateral sterile group, a four-celled carpogonial branch, and a one-celled basal sterile group. They also described a character found exclusively in Haraldiophyllum: the nuclei lie beneath the chloroplast (a feature observed for members in the tribe Cryptopleureae) while for other members of the Myriogrammeae the nuclei are positioned along the interior side walls of the cell (Lin et al. 2007). Additionally, Haraldiophyllum and Neoharaldiophyllum have carposporangia that are solitary and terminal, while carposporangia in Myriogramme are borne in terminal chains (Wynne 1983, Lin et al. 2007).
The Hawaiian species of Haraldiophyllum described here is clearly a member of the Myriogrammeae in terms of molecular phylogeny. No characteristics of reproductive development have been observed, which are considered diagnostic features for the group and the distinction from their known congeners, because no gametophytic plants were collected. Nonetheless, recognition of the species is supported by molecular phylogenetic data and comparisons of vegetative morphology. The molecular analyses included sequences representing the tribe Myriogrammeae (Figs 1 & 2), including the type species M. livida (J. D. Hooker & Harvey) and M. smithii (J. D. Hooker & Harvey) Kylin from the Falkland Islands and M. manginii (Gain) Skottsberg from the Antarctic Peninsula. The phylogenetic trees based on LSU and rbcL sequences revealed the same topology: within the Myriogrammeae, Myriogramme sequences formed a distinct clade that was sister to the clade containing all Haraldiophyllum and Neoharaldiophyllum species, with strong bootstrap and posterior probability support. These results are similar to those reported by Lin et al. (2007), Kim and Kang (2011), and Kang et al. (2017), confirming the position of the three genera in the Myriogrammeae and the tribal relationships within the subfamily Phycodryoideae. Interestingly, a third clade of Haraldiophyllum specimens from European waters (collected from private aquaria) identified as Haraldiophyllum sp. 2 was positioned as an early diverging lineage from Haraldiophyllum and Neoharaldiophyllum, with support, based on the rbcL tree (Fig. 1). The subfamily Myriogrammeae was clearly separated in two robust clades, with the Hawaiian specimens resolved as a basal taxon in the main Haraldiophyllum clade, grouping with H. bonnemaisonii, H. crispatum and Haraldiophyllum sp. 1 with high support, and supporting the conclusion that the Hawaiian specimens represent a distinct taxonomic entity from other congeners and also from Neoharaldiophyllum species. Two possible solutions could resolve the non-monophyly of Haraldiophyllum, given the phylogenetic position of Neoharaldiophyllum: (1) transfer all Neoharaldiophyllum species to Haraldiophyllum, making the genus monophyletic, or (2) recognize the different clades within Haraldiophyllum + Neoharaldiophyllum as a series of genera that include Haraldiophyllum sensu stricto. However, no morphological data are currently available for Haraldiophyllum sp. 1 and sp. 2 (vouchers MK125389, LT969700, LT969717, LT969714, and LT969716) (Supplementary Table S1) to aid in the delineation of these taxa. Additional investigation of morphological features for Haraldiophyllum species from Australia and Europe would be of great value in defining whether these specimens belong to this genus, or whether they represent a new genus to be described. Furthermore, a previous study already reported the necessity of additional morphological investigation to better define the genus: Lin et al. (2007) assessed H. bonnemaisonii, reported from Namibia and South Africa, and showed that it does not belong in Haraldiophyllum. Moreover, they demonstrated that H. sinuosum was established based on the description by Lucas (1913) of Nitophyllum sinuosum A. H. S. Lucas, and that it has not been reinvestigated in detail. To date there are no sequences for H. infossum and H. sinuosum, and phylogenetic analysis of these two species would be a valuable contribution toward understanding the diversity and evolutionary history of the group.
Many taxonomic investigations employing molecular phylogenies have considered the red algal female anatomy and post-fertilization structures as useful characters for defining generic boundaries in the Gigartinales, Halymeniales and Ceramiales (e.g., Lin et al. 2002, 2012, Díaz-Tapia et al. 2013, Gargiulo et al. 2013). Although classification based on the arrangement of carposporangia has been used to describe the Myriogrammeae, comparisons are only valid when all developmental stages are reported (Hommersand and Fredericq 1997). In the present study it was decided to classify our specimens as members of Haraldiophyllum rather than Neoharaldiophyllum, even though the molecular similarity of our specimens is greater with N. udoense than with H. bonnemaisonii and H. crispatum, due to the fact that for both molecular analyses, the Hawaiian specimens grouped with good support with other species of Haraldiophyllum, including the generitype H. bonnemaisonii.
In 2017, when the genus Neoharaldiophyllum was established, species of Haraldiophyllum were re-evaluated and two major changes were suggested: the Korean H. bonnemaisonii, described by Nam and Kang (2012) was found to be in agreement with the Neoharaldiophyllum-type, and Australian specimens of Neoharaldiophyllum nottii, as recorded by Womersley (2003, as Haraldiophyllum nottii) were found to be of the Haraldiophyllum-type and referable to H. crispatum (Kang et al. 2017), although the original material used to describe Neoharaldiophyllum nottii is from Washington, USA, and was recognized as Nitophyllum nottii by Norris and Wynne (1968) due to it presenting a Neoharaldiophyllum-type development of the cystocarp. Thus, the genus Haraldiophyllum has the following distribution: the type species H. bonnemaisonii has records for the Atlantic Ocean and the Mediterranean Sea, and H. crispatum has the broadest reported distribution including Australia and New Zealand, Chile, temperate South America, Antarctica and Subantarctic Islands. The remaining three species have more restricted distributions; H. sinuosum is known from Australia and Brazil, H. infossum has just one record in New South Wales, Australia, and H. hawaiiense is thus far known only from PMNM, Hawai‘i. Prior to this study, the depth range recorded for the genus was from shallow waters to 24 m (Kim and Kang 2011), but the specimens in this study were collected at depths between 81–93 m, making H. hawaiiense the only Haraldiophyllum species recorded from mesophotic depths thus far.
Haraldiophyllum hawaiiense is described here as a new species and as a new genus record from mesophotic depths in the Hawaiian Islands. Recent studies have characterized the algal biodiversity of both shallow and mesophotic environments, and based on a combination of molecular and morphological data, new species were described, including four new species of Ulvaceae (Spalding et al. 2016), two new species of Ramicrusta (Sherwood et al. in press), one new species of Sonderophycus (Sherwood et al. 2020), and four new species of Martensia (Sherwood et al. 2019). These are some of the first studies in an ongoing effort to use molecular and morphological data to characterize the mesophotic algal flora of the Hawaiian Islands. The biodiversity characterization of these deep-water environments will be essential to understand whether these MCEs serve as refugia for other organisms from shallow waters after experiencing stress, or are unique environments that merit recognition for their distinct diversity. Additionally, by exploring the endemism levels of species and the distinctiveness of shallow and deep floras, biogeographic comparisons will be possible with other mesophotic regions of the world.
SUPPLEMENTARY MATERIALSSupplementary Table S1Accession data for sequences used in phylogenetic analyses (https://www.e-algae.org). ACKNOWLEDGEMENTSWe thank the talented divers associated with the Papahānaumokuākea Marine National Monument, and the officers and crew of the NOAA ship Hi‘ialakai for access to the Hawaiian MCEs. This project was financially supported by the U.S. National Science Foundation (DEB-1754117), the U.S. National Fish & Wildlife Foundation (NFWF 0810.18.059023), and the National Oceanic and Atmospheric Administration (NOAA) Papahānaumokuākea Marine National Monument. The scientific views and conclusions, as well as any views or opinions expressed herein, are those of the authors and do not necessarily reflect the views of any of the above agencies.
Fig. 1Maximum likelihood phylogeny of rbcL sequences for Haraldiophyllum and closely related genera in the red algal subfamily Phycodryoideae. Numbers along branches indicate nodal support (first value, bootstrap support; second value, Bayesian posterior probabilities). Nodes with full support are indicated with an asterisk. Scale bar represents: substitutions per site. 
Fig. 2Maximum likelihood phylogeny from combined data of rbcL and LSU sequences for Haraldiophyllum and closely related genera in the red algal subfamily Phycodryoideae. Numbers along branches indicate nodal support (first value, bootstrap support; second value, Bayesian posterior probabilities). Nodes with full support are indicated with an asterisk. Scale bar represents: substitutions per site. 
Fig. 3Morphology of Haraldiophyllum hawaiiense sp. nov. (A) Live holotype specimen (BISH 776069), newly collected at 82 m depth at Salmon Bank Atoll, Hawai‘i (tetrasporic plant). Arrow shows where the blade was attached to a stone. (B) Habit of pressed specimen (BISH 776070). (C) Surface view of blade showing polygonal cells and the dentate margin (BISH 776071). (D) Surface view of the blade with tetrasporangial sori (BISH 776061). Arrows show elliptical tetrasporangial sori. (E) Cross-section of the blade showing the monostromatic structure. Arrows indicate pitconnections between cells (BISH 776069). (F) Cross-section of the polystromatic basal portion of a blade (BISH 776069). (G) Surface view of the basal portion of the blade showing the fan-shaped thickened nerve (BISH 776070). (H) Surface view of a tetrasporangial sorus (BISH 776069), with tetrasporangia (arrows). (I) A mature tetrasporangium, divided tetrahedrally (BISH 776069). (J) Cross-section of an immature tetrasporangial sorus. Arrows indicate immature tetrasporangia (BISH 776071). (K) Interior of a sorus, showing immature and mature tetrasporangia (BISH 776071). Scale bars represent: A & B, 1 cm; C, F, G & I–K, 100 μm; D, 250 mm; E, 50 μm; H, 250 μm. 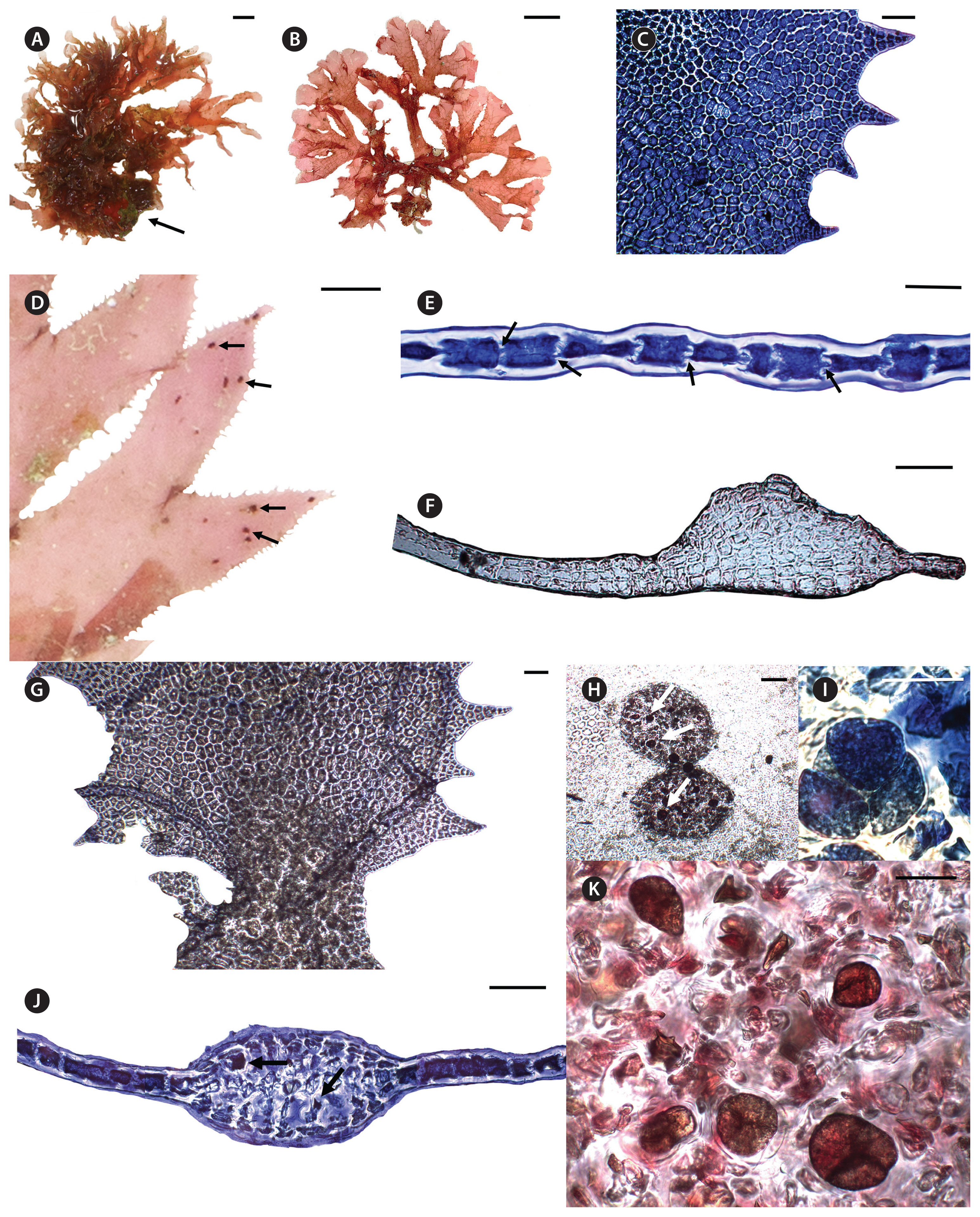
Table 1
Haraldiophyllum hawaiiense sp. nov. specimens from the Hawaiian Archipelago described in this study REFERENCESDíaz-Tapia, P., Boo, SM., Geraldino, PJL., Maneiro, I., Bárbara, I. & Hommersand, MH. 2013. Morphology and systematics of Calliblepharis hypneoides, sp. nov. (Cystocloniaceae, Rhodophyta) from the Atlantic Iberian Peninsula. Eur J Phycol. 48:380–397.
Edgar, RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 5:113 pp.
Gargiulo, GM., Morabito, M. & Manghisi, A. 2013. A re-assessment of reproductive anatomy and postfertilization development in the systematics of Grateloupia (Halymeniales, Rhodophyta). Cryptogam Algol. 34:3–35.
Gavio, B. & Fredericq, S. 2002.
Grateloupia turuturu (Halymeniaceae, Rhodophyta) is the correct name of the non-native species in the Atlantic known as Grateloupia doryphora
. Eur J Phycol. 37:349–359.
Guiry, MD. & Guiry, GM. 2020. AlgaeBase, World-wide electronic publication, National University of Ireland, Galway, Available from: https://www.algaebase.org
. Accessed May 1, 2020
Harper, JT. & Saunders, GW. 2001. Molecular systematics of the Florideophyceae (Rhodophyta) using nuclear large and small subunit rDNA sequence data. J Phycol. 37:1073–1082.
Hommersand, MH. & Fredericq, S. 1997. Characterization of Myriogramme livida, Myriogrammeae trib. nov. (Delesseriaceae, Rhodophyta). J Phycol. 33:106–121.
Huelsenbeck, JP. & Ronquist, F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
Kang, JC. & Kim, MS. 2013. A novel species Symphyocladia glabra sp. nov. (Rhodomelaceae, Rhodophyta) from Korea based on morphological and molecular analyses. Algae. 28:149–160.
Kang, JC., Yang, MY. & Kim, MS. 2017.
Neoharaldiophyllum, a new genus of Delesseriaceae (Rhodophyta) based on carposporophyte development and molecular data. Bot Mar. 60:515–532.
Kim, MS. & Kang, JC. 2011. A new Korean red algal species, Haraldiophyllum udoensis sp. nov. (Delesseriaceae, Rhodophyta). Algae. 26:211–219.
Kim, MS., Kim, SY. & Nelson, W. 2010.
Symphocladia lithophila sp. nov. (Rhodomelaceae, Ceramiales), a new Korean red algal species based on morphology and rbcL sequences. Bot Mar. 53:233–241.
Kylin, H. 1924. Studien über die Delesseriaceen. Lunds Univ. Årsskr. N F Avd. 20:1–111.
Kylin, H. 1925. The marine red algae in the vicinity of the Biological Station at Friday Harbor, Wash. Lunds Univ. Årsskr. N F Avd. 21:1–87.
Kylin, H. 1934. Bemerkungen fiber einige Nitophyllaceen. K Fysiograf Sdllskapets I Lund Fdrhandl. 4:1–10.
Lanfear, R., Calcott, B., Ho, SYW. & Guindon, S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
Lin, S., Hommersand, MH. & Nelson, WA. 2007. An assessment of Haraldiophyllum (Delesseriaceae, Rhodophyta), including H. crispatum (J.D. Hooker et Harvey) comb. nov. from New Zealand based on rbc L and LSU sequence analysis and morphological evidence. Eur J Phycol. 42:391–408.
Lin, S., Lewis, JE. & Fredericq, S. 2002.
Drachiella liaoii sp. nov., a new member of the Schizoserideae (Delesseriaceae, Rhodophyta) from Taiwan and the Philippines. Eur J Phycol. 37:93–102.
Lin, SM., Nelson, WA. & Hommersand, MH. 2012.
Hymenenopsis heterophylla gen. et sp. nov. (Delesseriaceae, Rhodophyta) from New Zealand, based on a red alga previously known as Hymenena palmata f. marginata sensu Kylin, with emphasis on its cystocarp development. Phycologia. 51:62–73.
Maggs, CA. & Hommersand, MH. 1993. Seaweeds of the British Isles. Vol. 1 Rhodophyta Part 3A. Ceramiales. HMSO, London, 444 pp.
Miller, MA., Pfeiffer, W. & Schwartz, T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE); Institute of Electrical and Electronics Engineers, Piscataway, NJ, 1–8.
Nam, KW. & Kang, PJ. 2012. Algal flora of Korea. Vol. 4 No. 7. Rhodophyta: Florideophyceae: Ceramiales: Delesseriaceae: 22 genera including Acrosorium. National Institute of Biological resources (NIBR), Incheon, 1–4.
Nam, KW. & Kim, YS. 1996.
Haraldiophyllum (Rhodophyta): a Delesseriaceous genus newly recorded in Korea. Algae. 11:101–106.
Norris, RE. & Wynne, MJ. 1968. Notes on marine algae of Washington and southern British Columbia, III. Syesis. 1:133–146.
Saunders, GW. 2005. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philos Trans R Soc Lond B Biol Sci. 360:1879–1888.
Sherwood, AR., Lin, S., Wade, RM., Spalding, HL., Smith, CM. & Kosaki, RK. 2019. Characterisation of Martensia (Delesseriaceae; Rhodophyta) from shallow and mesophotic habitats in the Hawaiian Islands: description of four new species. Eur J Phycol. 55:172–185.
Sherwood, AR., Paiano, MO., Spalding, HL. & Kosaki, RK. 2020. Biodiversity of Hawaiian Peyssonneliales (Rhodophyta): Sonderophycus copusii sp. nov., a new species from the Northwestern Hawaiian Islands. Algae. 35:145–155.
Sherwood, AR., Paiano, MO., Wade, RM., Cabrera, FC., Spalding, HL. & Kosaki, RK. Biodiversity of Hawaiian Peyssonneliales (Rhodophyta). 1. Two new species in the genus Ramicrusta from Lehua. Island Pac Sci. (in press).
Spalding, HL., Conklin, KY., Smith, CM., O’Kelly, CJ. & Sherwood, AR. 2016. New Ulvaceae (Ulvophyceae, Chlorophyta) from mesophotic ecosystems across the Hawaiian Archipelago. J Phycol. 52:40–53.
Stamatakis, A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
Womersley, HBS. 2003. The marine benthic flora of southern Australia. Part IIID: Ceramiales – Delesseriaceae, Sarcomeniaceae, Rhodomelaceae. Australian Biological Resources Study & the State Herbarium of South Australia, Canberra, 533 pp.
Wynne, MJ. 1983. The current status of genera in the Delesseriaceae (Rhodophyta). Bot Mar. 26:437–450.
Wynne, MJ. 2014‘2013’. The red algal families Delesseriaceae and Sarcomeniaceae. Koeltz Scientific Books, Königstein, 326 pp.
Zinova, AD. 1981. De positione systematica Nitophylli (Myriogrammes) yezoensis (Yamada et Tokida) Mikami (Delesseriaceae). Novit Syst Plant Non Vasc. 18:10–15.
|
|
|||||||||||||||||||||||||||||||||||||||||