Amazonocrinis thailandica sp. nov. (Nostocales, Cyanobacteria), a novel species of the previously monotypic Amazonocrinis genus from Thailand
Article information
Abstract
Cyanobacteria are distributed worldwide, and many new cyanobacterial species are discovered in tropical region. The Nostoc-like genus Amazonocrinis has been separated from the genus Nostoc based on polyphasic methods. However, species diversity within this genus remains poorly understood systematically because only one species (Amazonocrinis nigriterrae) has been described. In this study, two novel strains (NUACC02 and NUACC03) were isolated from moist rice field soil in Thailand. These two strains were characterized using a polyphasic approach, based on morphology, 16S rRNA phylogenetic analysis, internal transcribed spacer secondary structure and ecology. Phylogenetic analyses based on 16S rRNA gene sequences confirmed that the two novel strains formed a monophyletic clade related to the genus Amazonocrinis and were distant from the type species A. nigriterrae. The 16S rRNA gene sequence similarity (<98.1%) between novel strains and all other closely related taxa including the Amazonocrinis members exceeded the cutoff for species delimitation in bacteriology, reinforcing the presence of a new Amazonocrinis species. Furthermore, the novel strains possessed unique phenotypic characteristics such as the presence of the sheath, necridia-like cells, larger cell dimension and akinete cell arrangement in long-chains and the singularity of D1–D1’, Box-B, V2, and V3 secondary structures that distinguished them from other Amazonocrinis members. Considering all the results, we described our two strains as Amazonocrinis thailandica sp. nov. in accordance with the International Code of Nomenclature for Algae, Fungi and Plants.
INTRODUCTION
Cyanobacteria, as one of the earliest microorganisms, are a most important microbial group that contributes to oxygen production through photosynthesis and nutrient cycling (Brocke et al. 2015, 2018). They are commonly distributed in aquatic, terrestrial, and extreme environments (i.e., deserts, polar regions, hot springs, and endolithic habitats) (Whitton and Potts 2013, Tashyreva and Elster 2015, Bravakos et al. 2016, Strunecky et al. 2020, Dvořák et al. 2021). Cyanobacteria are the most morphologically diverse phyla of prokaryotes (Komárek 2016). Several studies have found that the morphological characteristics of some cyanobacteria do not reflect evolutionary relationships (Řeháková et al. 2007, Hašler et al. 2012, Shalygin et al. 2017, McGregor and Sendall 2021, Szubert et al. 2021). Recently, the polyphasic approach combining molecular, morphological, ecological and other data (ultrastructure, biochemical analysis) has been used for cyanobacterial classification to assist the recognition of new taxa (Komárek et al. 2014, Komárek and Johansen 2015, Komárek 2016). In modern cyanobacterial taxonomy, the 16S rRNA gene sequence data are utilized as molecular evidence that can differentiate cyanobacterial diversity at the genus level (Johansen and Casamatta 2005, Komárek and Mareš 2012). However, using 16S rRNA gene sequences alone for species-level identification may not have enough resolution, and other data are required for species evaluation (Dvořák et al. 2015). The 16S–23S rRNA internal transcribed spacer (ITS) region is also recommended for comparative analysis of secondary structures and offers information concerning divergence when the operational taxonomic units of studied strains are more closely related (Piccin-Santos et al. 2014). Thus, the polyphasic approach has become the basis for cyanobacterial taxonomic revision and the establishment of new taxonomic groups.
The cyanobacterial genus Nostoc, the type genus of the family Nostocaceae, has a complicated life cycle, with broad ecological diversity and insufficient morphological traits for differentiating this genus from closely related genera (Hrouzek et al. 2005, Řeháková et al. 2007, Papaefthimiou et al. 2008, Suradkar et al. 2017, Saraf et al. 2019). Furthermore, the genus Nostoc is polyphyletic in origin and a taxonomic revision of the genus is required (Hrouzek et al. 2005, Rajaniemi et al. 2005a, 2005b, Komárek et al. 2014). Based on the 16S rRNA gene phylogeny, one of the many clades for Nostoc-like strains was estimated as containing a representative of the type-species of the genus and defined as corresponding to the true Nostoc clade (also called the Nostoc sensu stricto clade) (Hrouzek et al. 2005, Řeháková et al. 2007, Papaefthimiou et al. 2008, Lukešová et al. 2009, Komárek et al. 2014). Thereafter, several Nostoc-like genera such as Aliinostoc, Compactonostoc, Desikacharya, Desmonostoc, Minunostoc, Mojavia, Halotia, Komareliella, Parakomarekiella, Pseudoaliinostoc, Purpureonostoc, Violetonostoc, Amazonocrinis, Atlanticothrix, and Dendronalium have been separated from the genus Nostoc by their distant position in phylogenetic analysis, morphological markers and other information including 16S–23S rRNA ITS sequences, ecological traits, genomics and in some cases even physicochemical characteristics in polyphasic approaches (Řeháková et al. 2007, Hrouzek et al. 2013, Genuário et al. 2015, Bagchi et al. 2017, Hentschke et al. 2017, Cai et al. 2019a, 2019b, 2020, Saraf et al. 2019, Soares et al. 2020, Alvarenga et al. 2021, Lee et al. 2021). The genus Amazonocrinis with type species A. nigriterrae was described based on the polyphasic study results of Nostoc-like strains from Brazil (Alvarenga et al. 2021). This genus is morphologically similar to Nostoc and has heterocytous filaments with uniseriate trichomes, without branching or meristematic zones, with single-pored heterocytes at just one end and two-pored heterocytes internally presented in trichomes. However, phylogenetic analysis based on the 16S rRNA gene and phylogenomic reconstruction of cyanobacteria genomes revealed that the genus Amazonocrinis was positioned distantly from the Nostoc sensu stricto (Alvarenga et al. 2021). Studies on Amazonocrinis mostly emanate from soil or freshwater samples collected in Brazil. Furthermore, species diversity within the genus Amazonocrinis is poorly understood systematically because only A. nigriterrae has been described.
Thailand is a tropical hotspot that contains unique biodiversity (Tantipisanuh et al. 2016) and novel cyanobacterial species have previously been described in this area (Komárek et al. 2010, Chatchawan et al. 2012, Kozlíková-Zapomělová et al. 2016, Tawong et al. 2019, 2022). This study presented the novel species within the genus Amazonocrinis from Thailand. Two novel Amazonocrinis strains isolated from a rice field in the central region of Thailand were characterized using a polyphasic approach. Based on their morphology, 16S rRNA gene sequence, 16S–23S rRNA ITS region secondary structures, the two novel strains presented dissimilar characteristics to the currently described species of the genus Amazonocrinis. Thus, we proposed the description of a novel Amazonocrinis species as Amazonocrinis thailandica sp. nov.
MATERIALS AND METHODS
Isolation and cultivation
A soil sample was collected from a rice field in Manorom District, Chai Nat Province, Thailand in June 2016. The sampling location was a rice field near the confluence of the Chao Phraya and Sakae Krang Rivers. The air temperature while sampling was 32°C. Aliquots of 0.1 mL of collected soil sample dilutions (103–105) were streaked into CT solid medium (Andersen 2005) following the slightly modification using NaNO3 (2,260 μmol L−1) as a nitrogen source until a single cyanobacterial colony appeared. Two strains that displayed Nostoc-like morphology were obtained and further cultured in screw-capped glass tubes containing 10 mL of modified liquid CT medium to investigate morphological features and life cycle. The strains were maintained at 25°C under light intensity of 30 μmol photons m−2 s−1 from white fluorescent lamps with a 12: 12 h (light: dark) cycle at the Naresuan University Algal and Cyanobacterial Collection, Naresuan University, Phitsanulok and labeled NUACC02 and NUACC03.
Morphological analyses
In both exponential and stationary phase cultures, morphological dimensions (e.g., length and width of the vegetative cells, heterocytes and akinetes) of the studied strains (NUACC02 and NUACC03) were investigated from >50 individuals using an optical photomicroscope BX53/DP72 (Olympus, Tokyo, Japan) with 400–1,000× magnification. The microphotographs were analyzed using an AxioVs40x64 v 4.9.10 imaging system (Carl Zeiss, Jena, Germany).
DNA extraction and polymerase chain reaction amplification
Unialgal cultures of strains used in this study were harvested and then rinsed at least three times in distilled water. To obtain the total genomic DNA, the rinsed pellet cell was extracted using the methods described previously by Tawong et al. (2019). Polymerase chain reaction (PCR) amplifications were conducted with the primer set 27F1/1494Rc (Neilan et al. 1997) for the 16S rRNA gene region and the primers 322F/340R (Iteman et al. 2000) for the 16S–23S rRNA ITS regions. The PCR reaction was performed in a MyCycler Thermal Cycler (Bio-Rad, Hercules, CA, USA). The amplification program was carried out as follows: one cycle of 2 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C and then a final 5 min elongation step at 72°C. The PCR products were purified using PureDireX PCR Clean-Up & Gel Extraction Kit (Bio-Helix, Keelung, Taiwan) following the manufacturer’s instructions. The purified PCR products were then sequenced using a BigDye Terminator v3.1 Cycle Sequencing Kit on an ABI PRISM 3730XL DNA analyzer (both from Applied Biosystems, Foster City, CA, USA). All primer sets used in the amplification were used for sequencing, while additional primers CYA359F, 809R, and 740F were also used for the 16S rRNA gene sequencing, as mentioned in Tawong et al. (2019). The nucleotide sequences of each primer set were assembled into contigs with 100% overlapping regions, and then the obtained consensus sequence was used for molecular analyses. All sequences in this study were deposited in GenBank under accession numbers LC657918–LC657919 for the 16S rRNA gene sequences and LC657920–LC657921 for the 16S–23S rRNA ITS region sequences.
Phylogenetic analyses
For phylogenetic analyses, the novel 16S rRNA gene sequences obtained in this study were aligned with the reference sequences from the GenBank database based on a BLAST search and previous studies (i.e., Cai et al. 2019a, 2019b, 2020, Alvarenga et al. 2021) using a MAFFT 7.309 (Katoh and Standley 2013). The GTR + G + I as the best-fit model of DNA nucleotide substitution based on the Akaike Information Criterion in MrModeltest v.2.4 (Nylander 2004) was obtained for the 16S rRNA gene dataset. The final phylogenetic analyses were conducted using maximum likelihood (ML) and Bayesian inference (BI) methods. The ML analysis with 1,000 bootstrap replications was run via RAxML 8.2.9 (Stamatakis 2014). The BI analysis was performed using MrBayes 3.2.7a (Ronquist et al. 2012). Two runs of four Markov chains were executed for 2.0 × 107 generations, sampling every 100th generation. In the BI analysis, the convergence of the runs was assessed using the average standard deviation of split frequencies, potential scale reduction factor (PSRF) and effective sample size (ESS) of all parameters in MrBayes 3.2.7a. The final average standard deviation of split frequencies below 0.01, a value of PSRF close to 1.00 and ESS values of all parameters above 200 were considered as good indicators of convergence. A 25% generation of each run was then discarded as burn-in and the remaining trees were used to calculate a 50% majority-rule consensus tree as well as posterior probabilities. For 16S rRNA gene sequences, genetic distance values (p-distance) were estimated using Mega X (Kumar et al. 2018) and then similarity values [100 × (1 - p)] were also calculated.
16S–23S rRNA ITS structure folding
Secondary structures of the ITS regions of our strains and their related phylogenetical taxa were predicted based on sequence regions of D1–D1’, V2, Box-B, and V3 helices. The putative regions found in the sequence were determined using RNA Folding Form V2.3 in the UNAFold (Markham and Zuker 2008).
Pigment spectral characterization
Phycobiliproteins as water-soluble protein pigments were extracted from cultures in 2 mL of 0.01 M potassium phosphate buffer pH 7.0, following Sciuto et al. (2017). The harvested cells were also extracted in 2 mL of methanol to obtain methanol-soluble pigments following Rastogi and Incharoensakdi (2014). Then, all samples were disrupted by freezing (−20°C) for 24 h and thawing at room temperature for 24 h. Cell debris was removed by centrifugation at 4°C and 10,000 ×g for 10 min. All absorbance spectra of each sample were recorded using a UV-1800PC Spectrophotometer (Shimadzu Corp., Kyoto, Japan) in the range 250–750 nm. Absorption spectra of the two different extracts were compared with the reference information described by Pagels et al. (2019) and Roshan et al. (2015).
RESULTS
Amazonocrinis thailandica Tawong, Pongcharoen, Pongpadung, Ponza & Saijuntha sp. nov. (Fig. 1)

Micrographs of Amazonocrinis thailandica. (A) Filament agglomerate. (B) Filament in mucilaginous sheath (s). (C–F) Filament characteristics showing barrel-shaped vegetative cells interleaved with purple cells. (G–I) Akinete cells (ak). (J) Single-pored heterocyte (sh) in terminal position. (K & L) Two-pored heterocytes (th) in intercalary or terminal position. (M) Single-pored heterocyte (sh) originating from vegetative cells adjacent to both sides of dying or releasing two-pored heterocytes cells. (N & O) Necrotic cells (nc) similar to disc or necridia present in the trichomes. (P) Cell division (cd) forming larger spherical colonies. (Q) New trichome (nf) releasing from akinete cells. (R & S) Cell division (cd) forming elongated trichomes surrounded with mucilage layer. (T) Hormogonia. Scale bars represent: A, 50 μm; B–T, 10 μm.
Description
Macroscopic colonies on solid agar are brown and amorphous with a smooth surface enclosed by mucilage. In liquid cultivation, they grow as gelatinous biomass on the bottom of the test tube or attached to the tube walls as irregular clusters enclosed by diffluent mucilage. The filaments are uniseriate, densely or loosely agglomerated in the diffluent mucilaginous matter, irregularly flexuous with akinete-like cells, blue-green to brown-reddish, sometimes interleave with purple cell. The sheath surrounding the filaments is mucilaginous, hyaline, and thick or thin, sometimes absent. The trichomes are cylindrical, flexuous and distinctly constricted at cross-walls. Vegetative cells are mostly barrel-shaped to spherical, blue-green to brown, granular in the central region, with distinct chromoplasm and centroplasm, with dimensions of 3.33–9.38 (mean 6.00) μm in length and 3.68–7.05 (5.12) μm in width with, 0.59–2.26 (1.17) length/width ratio. Heterocytes are terminal and intercalary, blue-green, solitary, very rarely in pairs, variable in shape and size as oblong to spherical and irregularly compressed in flexuous filament or old trichome. Single-pored heterocytes are usually isolated, mostly at the terminal or basal areas and originating from the cells at both sides of dying or releasing two-pored heterocytes cells, with dimensions 3.68–10.72 (6.49) μm in length and 3.80–7.76 (5.57) μm in width with 0.74–2.00 (1.17) length/width ratio. Two-pored heterocytes are mostly intercalary, 4.90–10.28 (7.16) μm in length, 4.53–7.15 (5.91) μm in width and 0.80–1.96 (1.23) length/width ratio. Akinetes are produced solitary or apoheterocytically in long-chains, intercalary or apical in position with a smooth surface, mostly spherical or subspherical, irregularly compressed in compact old trichomes, bright green or purple, showing high granulation in young akinete cells that developed from vegetative cells in trichomes, larger than the vegetative cells at 4.79–11.58 (7.92) μm length, 4.76–9.92 (7.15) μm width and 0.64–2.24 (1.14) in length/width ratio. Necrotic cells are similar to disc or necridia cells present in the filament. Cell divisions are in one plane forming uniseriate and elongated trichomes surrounded with mucilage layer or forming larger spherical and encapsulated colonies. Reproduction is by germination of akinete cells with releasing new trichomes and by hormogonia, formed by fragmentation at the junction of the necrotic cell. Hormogonia are common, straight, with rounded or irregularly compressed cells.
Diagnosis
Species are delimited by strong molecular and phylogenetic analyses using 16S rRNA gene sequences. The morphology differs from the type species A. nigriterrae and Amazonocrinis-related cyanobacteria by having large vegetative cells and hecterocytes, apoheterocytical akinetes and purple vegetative cells inserted in the trichomes with the presence of sheath and necridia-like cells. The distinctive structures of D1–D1’, V2, Box-B, and V3 helices from the 16S–23S rRNA ITS region sequence are also evidence for establishment as a new species of the genus Amazonocrinis.
Holotype (here designated)
A formaldehyde-fixed specimen of strain NUACC02 was deposited at the Queen Sirikit Botanic Garden Herbarium (QBG), the Botanical Garden Organization, Chiang Mai, Thailand under the designation QBG No. 132169.
Reference strain
Amazonocrinis thailandica NUACC02, deposited at the Thailand Institute of Scientific and Technological Research (TISTR) Culture collection, Thailand under the designation TISTR 9505.
Type locality
Found in moist rice field soil 500 m from Chao Phraya River (15°18′33.6″ N, 100°05′12.8″ E), Manorom District, Chai Nat Province, Central Thailand in June 2016 by Tawong, W.
Etymology
From Latin thailandica as the modern name of Thailand related to the location where the new species of the genus Amazonocrinis was isolated.
Gene sequences
LC657918 for the 16S rRNA gene region and LC657920 for the 16S–23S rRNA ITS region.
Phylogenetic and molecular analyses
Phylogenetic analyses based on the 16S rRNA gene sequences (1,394 bp) from 211 cyanobacterial taxa including A. thailandica strains (NUACC02 and NUACC03) were performed using the BI and ML methods. Because the BI and ML trees have the same topology, only the ML tree is shown in Fig. 2 and Supplementary Fig. S1. The new phylogenetic analyses based on the 16S rRNA gene sequences showed that two A. thailandica strains were grouped within the genus Amazonocrinis clade. In this genus clade, five subclades (A-I to A-V) were distinctly subdivided. The first subclade A-I formed a robust clade containing the type species A. nigriterrae CENA 67 (MW326972 and MN551902) (BI = 1.00 and ML = 100). This subclade was a sister clade with the second subclade A-II formed by a single strain A. nigriterrae CENA69 (MW326026). The third subclade A-III consisted of A. nigriterrae CENA18 (AY218827) and CENA238 (MN551905). The A. thailandica strains formed a distant and distinct clade as the subclade A-V with robustly supported values from BI (1.00) and ML (100) methods (Fig. 2, Supplementary Fig. S1). The subclade A-IV containing A. nigriterrae CENA66 (MW327025) formed as the earlier divergent species of the genus Amazonocrinis clade.

Maximum likelihood (ML) tree based on 16S rRNA gene sequences showing the phylogenetic position of Amazonocrinis thailandica. Numbers at nodes indicate posterior probability (>0.50) and bootstrap values (>50%) obtained from Bayesian inference and ML analyses, respectively. OTU, operational taxonomic unit. Black bars indicate posterior probability of 1.0 and bootstrap values of 100% at the same node. Bold font indicates the strain sequences studied.
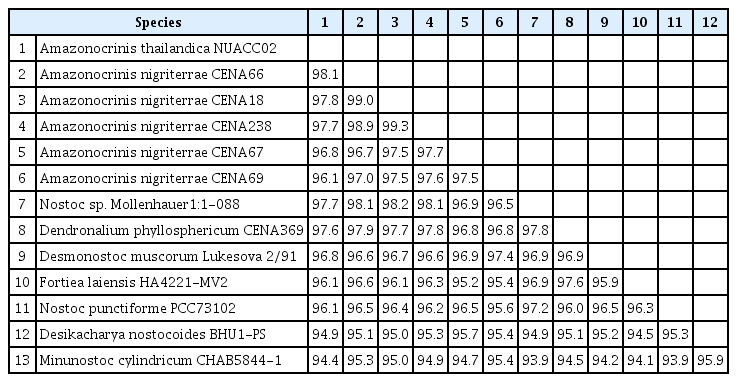
The evolutionary distance matrix based on the 16S rRNA gene sequences of two A. thailandica strains shared 99.7% similarity with each other, 96.1–98.1% similarity with other members of the Amazonocrinis clade (i.e., A. nigriterrae strains CENA66, CENA18, CENA238, CENA67, and CENA238), 96.1% similarity with Nostoc punctiforme PCC73102, and 94.4–97.7% similarity with the representative strains of other Nostoc-like genera (Table 1).
Characterization of ITS secondary structures
The complete ITS region between 16S and 23S rRNA genes of A. thailandica was sequenced and compared with other phylogenetically related taxa for which ITS sequences were available. In this study, only one operon (782 bp) was obtained from both novel strains. Based on Iteman et al. (2000), we identified typical sequence blocks, including conserved domains (D1–D5), major variable stems (V2 and V3), antiterminator (Box-B and Box-A). Furthermore, the 16S–23S rRNA ITS region sequences of A. thailandica strains also contained both isoleucine and alanine tRNAs. Putative secondary structures of D1–D1’, V2, Box-B, and V3 helices in A. thailandica were different and presented unique structures compared with the other strains (Fig. 3A–P). The D1–D1’ helix structure of A. thailandica was composed of a 5-bp helix basal stem (5′-AGGGA-UUCCG-3′), followed by a single base right bulge, a 6: 1 bp bilateral bulge, a 2 base left bulge, a 4: 5 bp internal loop and a 6 bp terminal hairpin loop (Fig. 3A). The D1–D1’ helix structures of A. thailandica and A. nigriterrae CENA 67 were almost identical but differed in the shape of the terminal hairpin loop. The D1–D1’ helix structure of A. nigriterrae CENA18 was very similar to A. nigriterrae CENA238 but slightly differed in the size of the terminal hairpin loop (Fig. 3C & D). The Box-B helix of A. thailandica was comprised of a 5-bp helix (5′-GGGAU-AUCUC-3′), followed by two large internal loops (3: 2 bp and 3: 3 bp, respectively) and a terminal hairpin loop with 10 bases (Fig. 3E). Besides differences in nucleotide length, the Box-B structure of A. thailandica, particularly the 3: 2 bp large internal loop and terminal hairpin loop, showed unique characteristics compared to Amazonocrinis clade members (Fig. 3E–H). The V2 helix of A. thailandica consisted of a 6 bp helix in the base of the stem (5′-GUAAUU-AAUUAU-3′) followed by 1: 1 and 2: 2 bp internal loops, as well as the terminal hairpin loop, contained 6 bp, which was very different from other phylogenetically related taxa (Fig. 3I). The V2-helix structures of the two A. nigriterrae strains CENA18 and CENA238 belonging to the Amazonocrinis clade were identical. The V2-helix structure of A. thailandica was also very different from Amazonocrinis members including A. nigriterrae CENA67, CENA18, and CENA238 (Fig. 3I–L). The V3 helix structure of A. thailandica was also distinct from all the other taxa (Fig. 3M–P). In detail, the V3 helix of A. thailandica consisted of very short stems (5′-GC-GU–3′), followed by a 1: 1 bp internal loop and further followed by a terminal hairpin loop with 5 bases.

Reconstruction of secondary structures of D1–D1’, Box-B, V2, and V3 helices within the 16S–23S rRNA internal transcribed spacer (ITS) region of Amazonocrinis thailandica and related taxa: (A, E, I & M) Amazonocrinis thailandica NUACC02. (B, F, J & N) Amazonocrinis nigriterrae CENA67. (C, G, K & O) Amazonocrinis nigriterrae CENA18. (D, H, L & P) Amazonocrinis nigriterrae CENA238. Gray highlights indicate similar structures among Amazonocrinis thailandica (A) and other members of the genus Amazonocrinis (B–D) for which ITS sequence data are available.
Pigment characterization
In the water-soluble extract, A. thailandica contained phycoerythrin as the dominant pigment with a high spectrum peak at 550 nm (Fig. 4A). In the methanolic extract, our strains showed general chlorophyll (436 and 665 nm) and carotenoid (473 nm) and also a peak indicating the likely presence of ultraviolet sunscreen pigment such as mycosporine-like amino acids at 337 nm (Fig 4B).
DISCUSSION
This study was carried out to ascertain the taxonomic status of novel Amazonocrinis species, named Amazonocrinis thailandica, from Thailand using an extensive polyphasic approach. Fiore et al. (2005) first indicated the possibility that an unknown clade containing Nostoc muscorum CENA18 (currently named as Amazonocrinis nigriterrae CENA18 in this study) would undergo reclassification based on its phylogenetic position. The taxonomic status of this clade was questionable because all contained strains were identified as Nostoc species. This unknown clade was later designated as the genus Amazonocrinis, and all Brazilian strains in this genus were described and named A. nigriterrae as a single species (Alvarenga et al. 2021). However, new phylogenetic analyses based on 16S rRNA gene sequences in this study revealed genetic divergence within the genus Amazonocrinis, with five distinct subclades (A-I to A-V). It has now been accepted that cyanobacterial species / genera exist as monophyletic clusters in the newest cyanobacterial taxonomy (Komárek et al. 2014, Dvořák et al. 2015, Komárek 2016, Johansen et al. 2017, 2021, Saraf et al. 2019, Alvarenga et al. 2021). Corresponding to the descriptions above, our results, confirmed by the 16S rRNA gene trees, indicated that A. thailandica formed a monophyletic clade that was distant from the type species A. nigriterrae CENA67. Findings suggested that A. thailandica had unique genetic traits and could be considered as a new species within the genus Amazonocrinis. Moreover, our 16S rRNA gene phylogenetic analyses corresponded to previous studies presenting that some sequences identified as Nostoc sp. also formed as a monophyletic clade (named as an unknown Nostoc-like clade in this study) (Fig. 2, Supplementary Fig. S1); however, this clade has not been investigated to define its taxonomic status (Fiore et al. 2007, Genuário et al. 2007).
The 16S rRNA gene similarity value has been utilized to represent evidence for separate genera or species of prokaryotic organisms including bacteria and cyanobacteria. Several authors suggested that the 16S rRNA gene sequence similarity considered for species delimitation should be below 98.7% (Stackebrandt and Ebers 2006, Kim et al. 2014, Yarza et al. 2014). Later, a <99% 16S rRNA gene similarity cutoff values was suggested to delimit species for nostocalean strains (Kaštovský et al. 2014). In this study, A. thailandica shared <98.1% 16S rRNA gene similarity with the closest phylogenetical taxa within the Amazonocrinis clade, and less than the species cutoff proposed by Stackebrandt and Ebers (2006), Kim et al. (2014), Yarza et al. (2014), and Kaštovský et al. (2014) (Table 1). This provided strong evidence supporting a species separation of A. thailandica from all other Amazonocrinis members. Therefore, the evolutionary distance based on the 16S rRNA gene sequence provided molecular evidence supporting the existence of A. thailandica as a new species of the genus Amazonocrinis.
Additional molecular data from the utilization of ITS secondary structures together with the 16S rRNA gene sequences have been advocated as essential for the recognition of cyanobacterial species (Osorio-Santos et al. 2014, Mai et al. 2018, Cai et al. 2021, Pietrasiak et al. 2021). In particular, the D1–D1’, Box-B, V2, and V3 helices in the ITS region between 16S–23S rRNA sequences are important for the separation of strains in different species (Johansen et al. 2011). Noticeably, among Amazonocrinis strains, the presence of a large bilateral bulge in the D1–D1’ and the middle bilateral bulge in Box-B were found to be consistent in all four strains studied (Fig. 3A–H). These structures may be unique characteristics of the genus Amazonocrinis and can be used to confirm the correction of ITS sequence data. The ITS secondary folding patterns of A. thailandica were significantly different from other Amazonocrinis taxa and closely related taxa within Nostocaceae reported in previous studies (Osorio-Santos et al. 2014, Mai et al. 2018, Alvarenga et al. 2021, Cai et al. 2021, Pietrasiak et al. 2021), supporting the establishment of this novel Amazonocrinis species. A. thailandica showed distinctly different structures of the V2 and V3 helices from all other Amazonocrinis taxa, highlighting the novelty of our species and supporting the phylogenetic analyses (Fig. 3I–P). Furthermore, the ITS secondary structures exhibited different folding patterns between A. nigriterrae strains CENA18, CENA238, and CENA67 indicating a separate species (Fig. 3). When considering the phylogenetic position and lower than the species-level threshold (<98.7%) of 16S rRNA gene sequence similarity (Kaštovský et al. 2014, Yarza et al. 2014), the four strains CENA 69, CEN18, CENA238, and CENA66 may bedifferent species within the genus Amazonocrinis. Thus, we expect that other Amazonocrinis species could be discovered when extra morphological and molecular evidence is available. Increased sampling within the putative new species (A-II to A-IV) would help to observe and identify the divergence within this group.
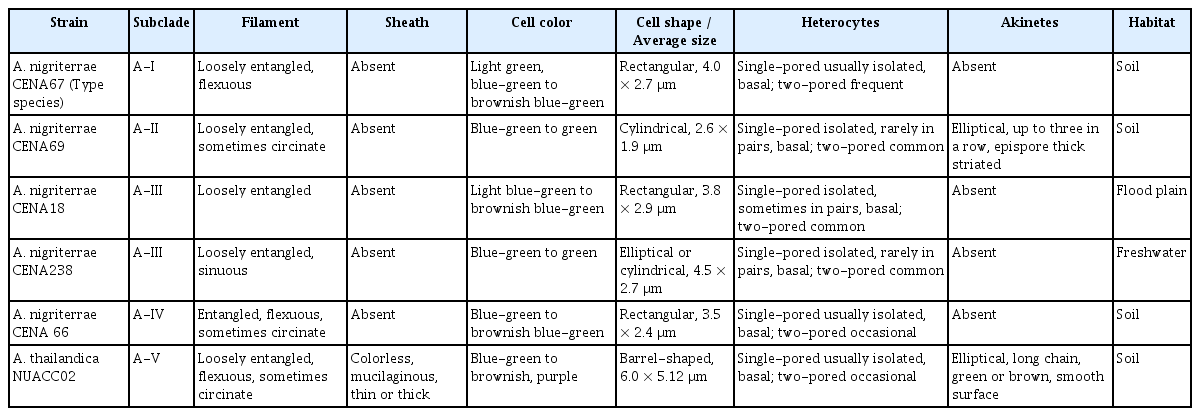
Morphologically, A. thailandica is very similar to Nostoc sensu stricto and Nostoc-like genera including Desmonostoc, Aliinostoc, Minunostoc, Halotia, and Komarekiella, because of the overlapping features in all these genera. For example, A. thailandica presents diffluent mucilaginous colonies similar to Minunostoc, Halotia, and Komarekiella that differ from typical Nostoc, Aliinostoc, Desmonostoc, and Mojavia. Furthermore, cell division in A. thailandica occurs by dividing into one plane in trichomes resembling Nostoc and Mojavia, whereas Halotia and Komarekiella divide into two planes. A. thailandica and Minunostoc follow the same type of cell division in trichomes with colonies wrapped by a diffluent mucilaginous sheath but Minunostoc does not produce heterocytes and akinete cells throughout the life cycle. Furthermore, our new species also present necrotic cells resembling discs or necridia that may relate to sporogenous cycle stage of Nostoc (Caiola and Pellegrini 1979). Differences in distinguishing A. thailandica from the type species Amazonocrinis and Amazonocrinis-like cyanobacteria include the presence of distinctly large cell dimensions in vegetative cells, purple vegetative cells inserted in trichomes, diffluent mucilaginous sheaths and the presence of large akinete cells (Table 2).
Results of pigment spectra analyses were also in accordance with the type species of Amazonocrinis showing purple or brown filaments containing Pe pigment (Alvarenga et al. 2021). Furthermore, the Pe found in our species is a valuable compound that can be applied in various industries such as aquaculture, food, cosmetics, medicine, biotechnology, and pharmacology (Pagels et al. 2019). However, the quality and quantity of light play a vital role in growth and pigment accumulation in cyanobacteria that adapt to different light environments as complementary chromatic adaptation (Hirose et al. 2019). Thus, light influence on phenotype color should be determined.
In terms of ecological information, most members in the genus Amazonocrinis occur in soil or sediments, except for A. nigriterrae CENA238 which is found in freshwater tropical areas of Brazil. In our study, new species of this genus were found in a rice field in Thailand more than 15,000 km from Brazil. This indicated that the genus Amazonocrinis might be found in different tropical zone areas. However, the observed morphological characteristics are insufficient and comparisons between A. thailandica and the other members of the genus Amazonocrinis remain difficult. Therefore, molecular evidence is an important criterion for distinguishing separate species.
Results revealed intra-species genetic diversity in the genus Amazonocrinis. A novel species called A. thailandica sp. nov. was described based on molecular and morphological evidence. However, cyanobacterial diversity in tropical regions requires further detailed investigation. Future studies using the polyphasic approach are necessary, with increased sample numbers in this region.
ACKNOWLEDGEMENTS
This study was supported by Naresuan University, Thailand (R2565B004).
Notes
The authors declare that they have no potential conflicts of interest.
SUPPLEMENTARY MATERIALS
Uncollapsed maximum likelihood (ML) phylogenetic analysis based on 16S rRNA gene sequences, with all sequences used to construct the collapsed phylogeny shown in Fig. 2 (https://www.e-algae.org).