INTRODUCTION
Marine ecosystems worldwide are experiencing rapid environmental variation due to climate change, which affects the phylogeographic patterns and genetic diversity of marine species, including macroalgae (Pauls et al. 2013, Neiva et al. 2015, Pinsky et al. 2020). Intraspecific genetic diversity is a fundamental component of a species’ adaptive potential to climate change (Wernberg et al. 2018); therefore, it is important to examine the genetic diversity of populations across a species’ range (Pauls et al. 2013). In the northwestern Pacific, several phylogeographic studies on macroalgae have been conducted to understand species distribution and intraspecific genetic diversity; these studies concluded that current phylogeographic patterns is derived from a complex interplay of spatial and temporal variation, including demographic history, lineage interactions, and local adaptation to environment (Kim et al. 2012, 2014, Hu et al. 2015, 2016, Yang et al. 2020, 2021a, 2021c). As species distribution and intraspecific genetic diversity are strongly influenced by evolutionary processes, understanding how these processes contribute to contemporary patterns can provide valuable insights into the adaptive potential of species and their future persistence in a changing environment (Avise et al. 1987).
The genus Chondrus Stackhouse comprises perennial red seaweeds with economic value that occur on rocks in lower intertidal areas (Taylor and Chen 1994). The type species, C. crispus Stackhouse, has long been harvested as a source of carrageenan used as a thickener and gelling agent, and for fiber and minerals in foodstuffs (Lamont and McSweeney 2021). Although 101 species, including 66 infraspecific ranks, have been listed in AlgaeBase, only 11 species are accepted taxonomically based on the global algal database (Guiry and Guiry 2021). Chondrus species have usually been distinguished by their thallus shape and the position of reproductive structures (Okamura 1936, Mikami 1965); however, accurate taxonomic identification based on phenotypic traits is difficult owing to their morphological plasticity (Brodie et al. 1993). Phylogenetic analysis of the family Gigartinaceae based on the plastid ribulose 1,5-bisphosphate carboxylase oxygenase large subunit gene (rbcL) show that the genus Chondrus is monophyletic and has a terminal position in the family (Hommersand et al. 1999). Recent phylogeographic studies of the type species, C. crispus, distributed throughout the North Atlantic, indicate that its current geographic range resulted from an interplay between ancestral survival in the east Atlantic during the Pleistocene glaciations and contemporary trans-oceanographic gene flow to the west Atlantic (Hu et al. 2007, 2010, 2011, Provan et al. 2013).
The hot-spot of species diversity of Chondrus is the northwestern Pacific (Mikami 1965). Ten Chondrus species have been reported in algal flora of Korea (Nam and Kang 2015): C. armatus (Harvey) Okamura, C. canaliculatus (C. Agardh) Greville, C. crispus Stackhouse, C. elatus Holmes, C. giganteus Yendo, C. nipponicus Yendo, C. ocellatus Holmes, C. pinnulatus (Harvey) Okamura, C. verrucosus Mikami, and C. yendoi Yamada et Mikami. Of these, C. ocellatus was first described in Japan based on specimens from Shimoda with a wide distribution along the intertidal areas of Korea, China, and Japan (Yoshida and Yoshinaga 2010, Nam and Kang 2015). Okamura (1932, 1936) proposed five forms based on morphological characteristics: f. ocellatus, f. canaliculatus, f. giganteus, f. crispus, and f. nipponicus. Mikami (1965) later raised f. canaliculatus and f. giganteus to species rank. Based on the life history and morphological evidence, Brodie et al. (1991) placed f. crispus and f. nipponicus into the congeneric species C. nipponicus, which has simple blades with many marginal proliferations. The observations of Brodie et al. (1991) showed that C. nipponicus is not always dichotomously branched, but frequently has an irregular branching pattern.
Despite rich species diversity and considerable economic potential, the number of valid Chondrus species remains a matter of debate in the literature, and little relevant data are available on intraspecific genetic diversity and phylogeography for this genus in the northwest Pacific (Matsumoto and Shimada 2013, Hu et al. 2015). A previous study on the phylogeography of Chondrus illustrated the genetic structure and biogeographic distribution of C. ocellatus in the northwest Pacific (Hu et al. 2015); however, the extent of sampling was insufficient to cover the distribution range in Korea. Furthermore, molecular phylogenetic relationships among Chondrus species in Korea have not been well addressed, with only one paper on C. retortus Matsumoto et Shimada available to date (Kang et al. 2020).
In this study, we conducted molecular analyses of the three Chondrus species, C. nipponicus, C. ocellatus, and C. giganteus, using specimens collected in Korea and Japan and sequences from China in GenBank. Our major aims were to (1) assess the levels and patterns of intraspecific genetic diversity based on the 5′ end of the cytochrome oxidase subunit I (COI-5P) gene; (2) compare phylogeographic structures among the three Chondrus species; and (3) confirm the phylogenetic relationships of this important genus based on the plastid rbcL gene. This phylogeographic knowledge is useful and urgent for safeguarding these natural seaweed resources with high morphological plasticity and commercial applications.
MATERIALS AND METHODS
Sample collection
Chondrus specimens were collected from the intertidal zones throughout the coast of Korea and in several regions of Japan (Supplementary Tables S1–S3). The DNA sequences data from China (Lvshun [LS], XiaoShi Island [XS], Chang Island [YC], DaQing Island [DQ], DouLuan Island [DD], Li Island [LI], Zhu Island [ZD], GouJi Island [GI], Yinghai beach in Qingdao [YH], and BaDaGuan in Qingdao [BD]) and Japan (Awaji Island [AJ], Shikanoshima [SF], Oki District [OK], Kotsubo [KT], and Niigata [NG]) were obtained from an earlier study (Hu et al. 2015). The field-collected specimens were divided into at least three groups (C. ocellatus, C. nipponicus, and C. giganteus) based on the morphology described in the literature (Mikami 1965, Brodie et al. 1991, 1993). And then a 2–3 cm portion of the frond was excised from each plant and desiccated in silica gel for DNA analysis. Voucher specimens were deposited in the herbarium of Jeju National University (JNUB), Jeju, Korea. Voucher images of representative isolates of each species are presented in Figs 1–4.
DNA extraction and sequencing
Genomic DNA was extracted using a LaboPass Tissue Genomic DNA Isolation kit (Cosmogenetech, Seoul, Korea) or a MagPurix Plant DNA Extraction kit (Zinexts, Taipei, Taiwan). A 535-bp long portion of mitochondrial COI-5P gene was amplified using newly designed primers in this study, ChondrusF1 (TTTTCAGGTGTTTTAGGTG) and ChondrusR1 (TGCATCAAAGAAAGAAGTAT). Similarly, a 1,233-bp long portion of plastid rbcL gene was assembled by amplifying two overlapping fragments using primers F145 and R898 for the first fragment, and F762 and R1442 (Kim et al. 2010) for the second fragment. All polymerase chain reaction (PCR) amplifications were conducted in an All-In-One-Cycler (Bioneer, Daejeon, Korea) using Master Mix 2x (MGmed, Seoul, Korea), under the following conditions: for COI-5P, initial denaturation at 96°C for 4 min, 40 cycles of denaturation at 94°C for 30 s, annealing at 45°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 7 min; and for rbcL, 96°C for 4 min, 35 cycles of 94°C for 1 min, 50°C for 1 min, 72°C for 2 min, and 72°C for 7 min. All PCR runs included a negative control containing all reagents, except for template DNA. Successful PCR products were purified using an Exo-AP PCR Clean-up Mix (MGmed) and sequenced at Macrogen (Seoul, Korea). The forward and reverse strands were determined for all specimens. Raw sequence reads for each gene were edited and assembled in Geneious R9 ver. 9.1.4 (Biomatters, Auckland, New Zealand), and aligned using MUSCLE (Edgar 2004) in Geneious. Consensus sequences were aligned with reference sequences from Hu et al. (2015).
Phylogenetic and genetic diversity analyses
In total, the 40 rbcL sequences, including 20 published sequences, were used for phylogenetic analysis. The sequences from the type localities of three Chondrus species (C. nipponicus, C. ocellatus, and C. giganteus) in Japan were included (Hommersand et al. 1994, 1999, Matsumoto and Shimada 2013). Phylogenetic trees were constructed based on rbcL sequence data using maximum likelihood (ML) in RAxML v. 8.2.10 (Stamatakis 2014). The GTR + GAMMA model was applied to the phylogenetic analyses. Statistical support for the nodes under ML was assessed using 1,000 bootstrap replicates. As Chondrus has been resolved as a monophyletic group (Hommersand et al. 1999), two Mazzaella species (M. californica and M. japonica) belonging to the family Gigartinaceae were designated as outgroups.
To evaluate genetic diversification patterns in the three Chondrus species based on mitochondrial COI-5P, we measured the number of haplotypes (Nh), haplotype diversity (H), and nucleotide diversity (π) using ARLEQUIN 3.5 (Excoffier and Lischer 2010). Relationships among COI-5P haplotypes of Chondrus were assessed based on haplotype networks that were constructed using ARLEQUIN. Uncorrected p-distances implemented in MEGA X (Kumar et al. 2018) were used to calculate genetic divergence in terms of pairwise differences between and within species. Historical demography such as potential population growth and expansion were inferred from neutrality tests and Tajima’s D and Fu & Li’s FS (Tajima 1989, Fu 1997). Under the assumption of neutrality, negative values always characterize population expansion (Tajima 1989, Fu 1997).
RESULTS
Phylogenetic analyses using the rbcL gene
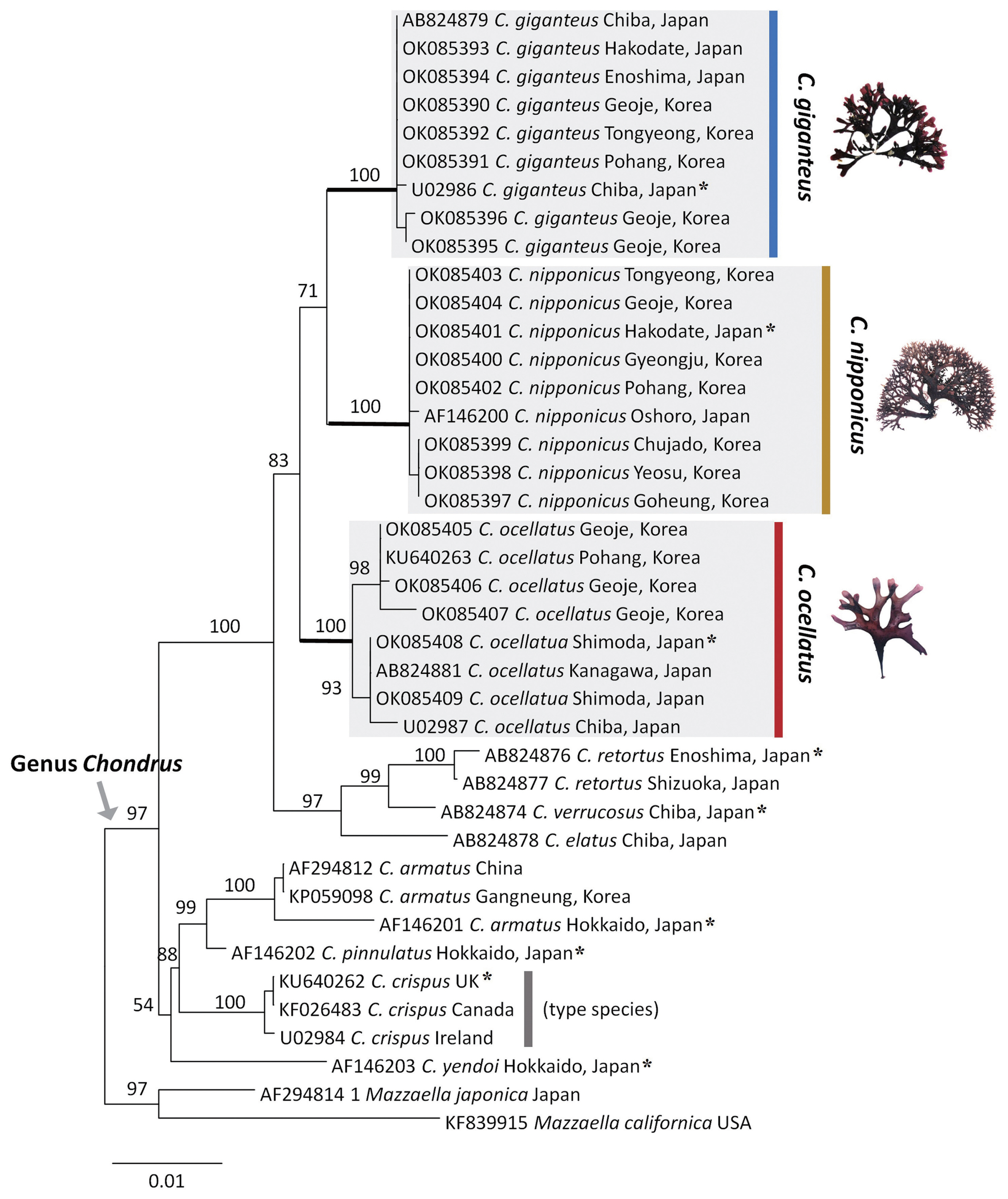
A ML phylogenetic tree based on rbcL yielded a well-resolved monophyletic clade in the genus Chondrus (Fig. 1). RbcL sequence alignment comprising 40 specimens (20 new sequences) and 1,233 nucleotides revealed 143 variable positions (12%) and 81 phylogenetically informative positions (7%). Newly generated rbcL sequences of Chondrus were presented in Supplementary Table S1. The specimens collected in this study were clearly divided into three clades: C. nipponicus, C. ocellatus, and C. giganteus (Fig. 1). They were strongly supported by 100% bootstrap values, including specimens from the type localities (AF146200, C. nipponicus from Oshoro in Japan; OK085408 and OK085409, C. ocellatus from Shimoda in Japan; AB824879 and U02986, C. giganteus from Chiba in Japan). These three species formed a sister clade to the following three species, C. verrucosus (AB824974), C. retortus (AB824876 and AB824877), and C. elatus (AB824878) from Japan. The occurrence of C. giganteus in Korea is a new report based on molecular evidence in this study.
Maximum intraspecific divergences were up to 0.16% for C. nipponicus, 0.97% for C. ocellatus, and 0.24% for C. giganteus. The interspecific divergences among the three species ranged from 1.30% (C. nipponicus and C. giganteus) to 2.03% (C. ocellatus and G. giganteus) (Table 1). The type species of Chondrus, C. crispus Stackhouse from the UK (KU640262), was distinct from our three species (C. nipponicus, C. ocellatus, and C. giganteus) and formed a sister clade to three other species, C. armatus (KF026483, AF294812, and KP059098), C. pinnulatus (AF146202), and C. yendoi (AF146203) from Japan.
Genetic diversity and phylogeographic structure using the COI-5P gene
The 777 Chondrus specimens based on interspecific differences using the COI-5P gene sequence were assigned as follows: 413 sequences of C. nipponicus, 225 of C. ocellatus, and 139 of C. giganteus (Table 2). The COI-5P sequences consisted of 535 positions and yielded 17, 13, and 2 polymorphic sites for C. nipponicus, C. ocellatus, and C. giganteus, respectively. The overall genetic diversity was similar between C. nipponicus (H = 0.5300, π = 0.0027) and C. ocellatus (H = 0.6256, π = 0.0034), but much lower for C. giganteus (H = 0.0563, π = 0.0002) (Table 2). The pairwise genetic distance within species ranged from 0.03% (C. giganteus) to 1.3% (both C. nipponicus and C. ocellatus) (Table 1). The interspecific divergence among the three species ranged from 3.3 to 5.6% (Table 1). We found different phylogeographic structures in each species and confirmed that branching patterns have variations without consistent characteristics even within the same species (Figs 2–4). The three species showed generally dichotomous branching with many marginal proliferations, except for C. giganteus.
For C. nipponicus, we detected 14 haplotypes, including two major haplotypes (CN1 and CN10), representing 89% of the specimens analyzed (Fig. 2). The major haplotype CN1 (n = 273) was distributed throughout Korea, China, and Japan (Fig. 2). Specimens from China and Japan were mostly assigned to CN1 (Supplementary Table S2). In Korea, CN1 was distributed from the southern to the eastern coasts, but not the western coast. The haplotype CN10 (n = 96) differed from CN1 by three mutations and was distributed only in Korea, from the western to the southern coasts. The COI-5P genetic diversity in the eastern and southern coast of Korea was generally higher than in the western coasts of Korea, China, Japan, and Jeju Island, Korea (Table 2).
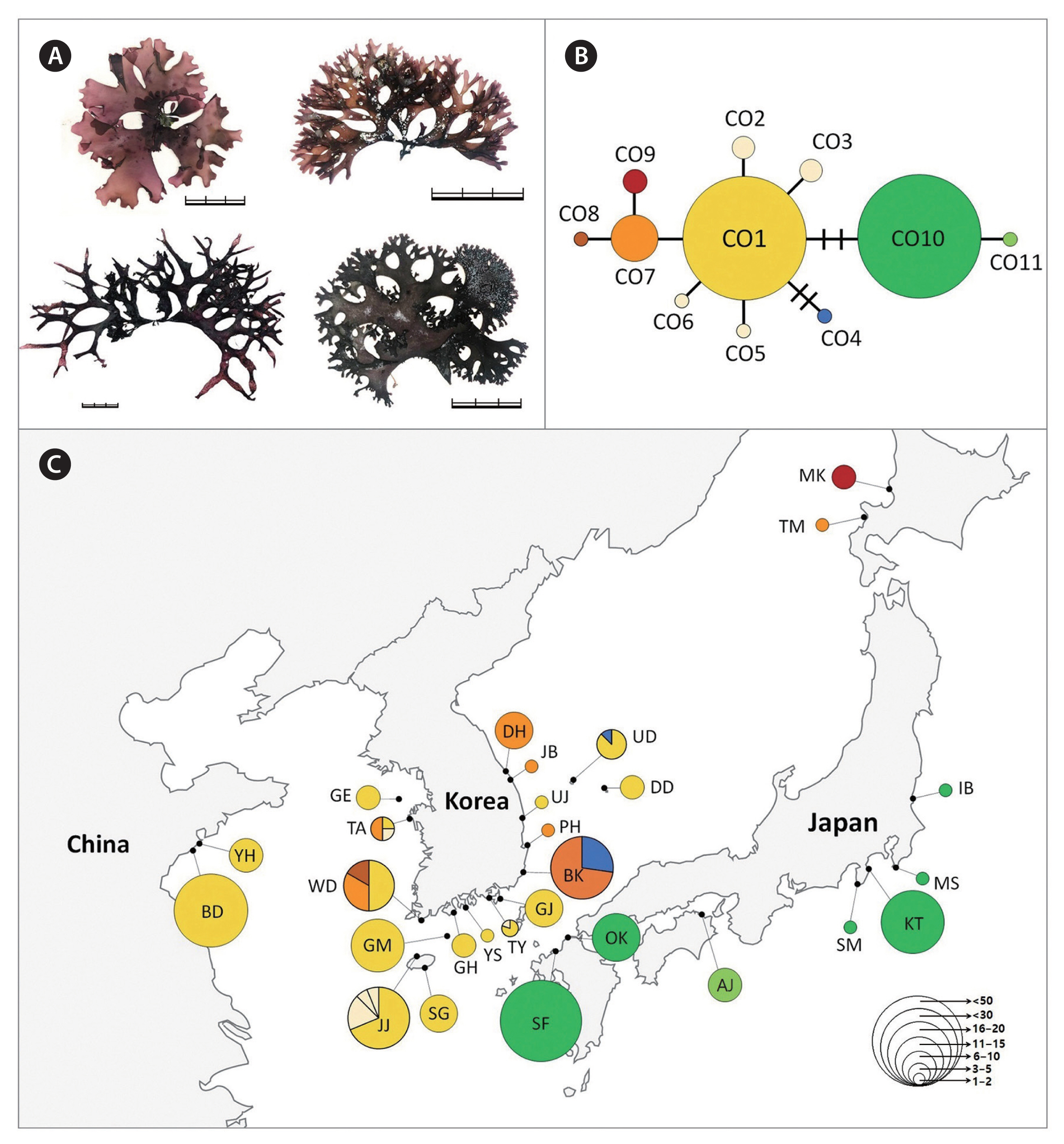
For C. ocellatus, we found 11 haplotypes, including two major haplotypes (CO1 and CO10), representing 80% of the specimens analyzed (Fig. 3). The major haplotype CO1 (n = 111) was widely distributed across the entire Korean coasts and Qingdao in China (BD, YH) (Supplementary Table S3). Several haplotypes (CO2–CO6) specifically occurring in Korea were derived from CO1, showing a star-shaped haplotype structure similar to C. nipponicus. The haplotype CO10 (n = 71) differed from CO1 by three mutations and occurred exclusively in southern Japan. Northern Japan (Tomari [TM], Mashike [MK]), having two haplotypes CO7 (TM) and CO9 (MK), exhibited a different haplotype distribution from the south. COI-5P data revealed higher genetic diversity in the western and southern coasts of Korea than in the eastern coasts of Korea, China, Japan, and Jeju Island (Table 2).
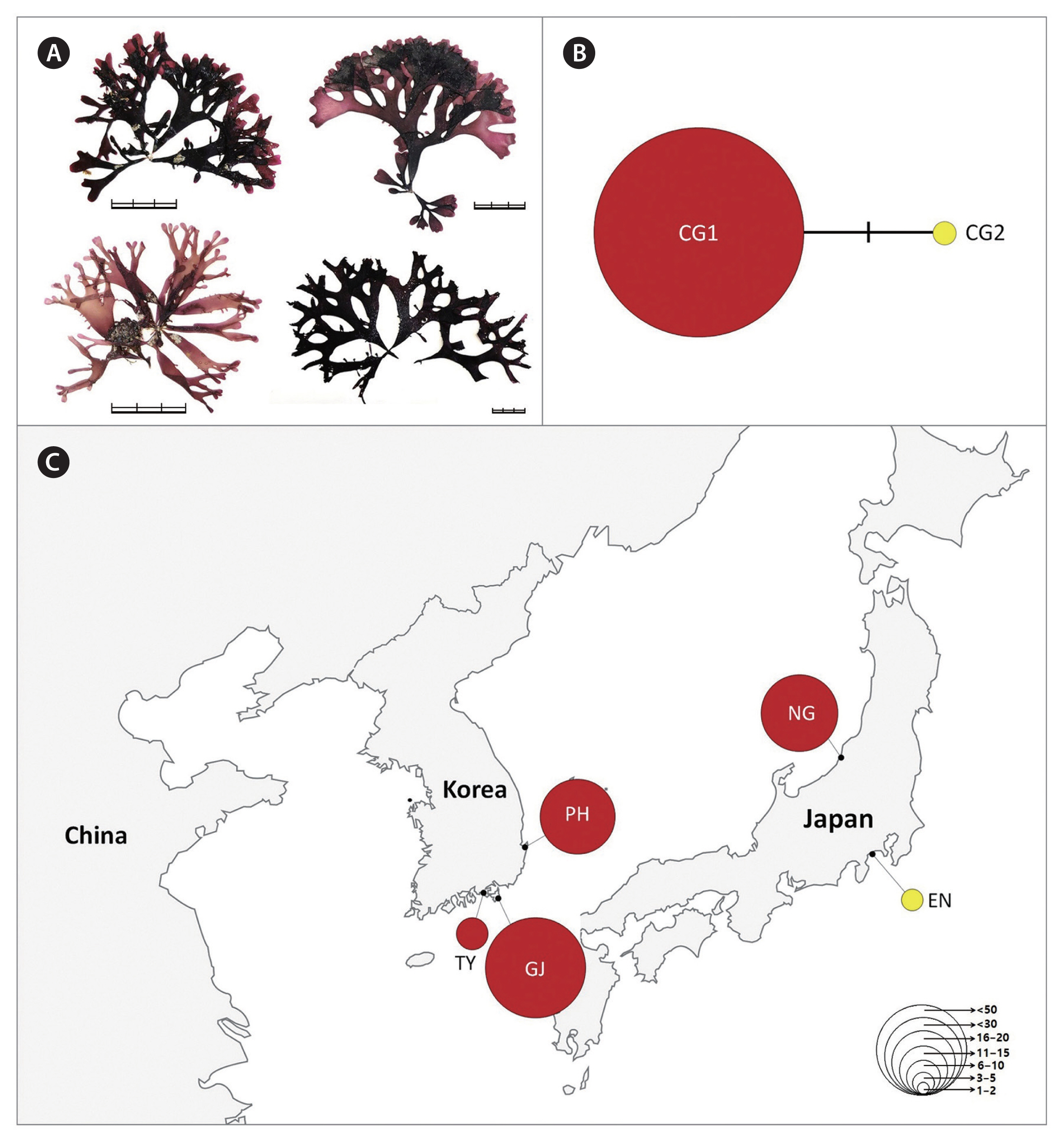
For C. giganteus, we defined two haplotypes separated by two mutations (Fig. 4). The major haplotype CG1 (n = 135) included all individuals from Korea and NG, Japan (Fig. 4, Supplementary Table S4). Specifically, a haplotype derived from CG2 was found in Enoshima (EN), Japan. C. giganteus had only a limited distribution in the southern coast of Korea (Pohang [PH], Geoje [GJ], Tongyoung [TY]), compared with the other two Chondrus species.
Neutrality tests indicated a recent population expansion for all three Chondrus species, with negative Tajima’s D and Fu’s FS indices (D = −1.051 and FS = −3.312 for C. nipponicus, D = −0.4061 and FS = −0.8298 for C. ocellatus, and D = −0.9994 and FS = −0.1730 for C. giganteus).
DISCUSSION
Chondrus species are important components of the coastal marine ecosystem in the northwest Pacific Ocean and contribute significantly to carrageenan production (Kamiya et al. 2021). However, the genetic diversity and phylogeographic structure of these economic red algae are topics still in need of study. The identification of Chondrus species is mainly based on morphological features, such as the branching pattern of the thallus and the position of reproductive structures (Mikami 1965, Kim 1976). Taxonomists have proposed several forms of Chondrus species from the northwest Pacific (Okamura 1932, 1936, Mikami 1965), despite drawbacks of morphology-based species delimitation. However, molecular techniques applied in other red algae have led to the discovery of multiple cryptic species that were previously classified as a single species (Yang and Kim 2018, Nauer et al. 2019, Yang et al. 2021a, Preuss et al. 2022). Simultaneously, multiple species defined by morphological traits were found to belong to a singular species using molecular data (Kang et al. 2015). Therefore, DNA-based phylogenetic and biogeographic analyses have become a well-appreciated method to assess the congruence between morphological and genetic variation of taxa (Furfaro et al. 2021, Gąsiorek et al. 2021).
In this study, we confirmed the phylogenetic relationships in the genus Chondrus, and compared genetic diversity patterns and phylogeographic structures among three Chondrus species in the northwest Pacific Ocean. Based on the phylogenetic results including topotypic specimens, we assigned the specimens to the well-resolved clades, C. giganteus, C. nipponicus, and C. ocellatus. Moderate intraspecific genetic diversity and the star-shaped haplotype network show that C. nipponicus and C. ocellatus share similar phylogeographical patterns, whereas C. giganteus shows extremely low intraspecific genetic diversity.
Few studies on the genus Chondrus have documented interspecific phylogeographic structures with topographical barriers in wide ranges that restrict gene flow and drive genetic divergence (Hu et al. 2011, 2015). The phylogeography and demographic history of C. ocellatus in the northwest Pacific Ocean revealed three clearly separated lineages (lineages A, B, and C) (Hu et al. 2015). These deep divergences in C. ocellatus appeared to have resulted from ancient isolation accelerated by random drift and limited genetic exchange between regions. In comparison with these findings, however, we found that the three species in our study, C. ocellatus, C. nipponicus, and C. giganteus, exactly matched with the lineages A, B, and C defined by Hu et al. (2015). This implies that the three Chondrus species were recognized as one species, C. ocellatus, in the study by Hu et al. (2015). As the identity of the three species has been confirmed by other phycologists (Brodie et al. 1991, 1993, 1994), the three cryptic lineages within C. ocellatus suggested by Hu et al. (2015) might be a misidentification caused by the morphological plasticity mentioned above.
Comparison of geographic patterns of intraspecific traits provides a clue into population divergence within species (Arbeláez-Cortés et al. 2014). MtDNA COI-5P gene sequencing shows that C. ocellatus and C. nipponicus have certain subtle regional differences in genetic diversity (Table 2). C. nipponicus exhibits a high genetic diversity in the eastern (H = 0.7524, π = 0.0031) and southern (H = 0.5555, π = 0.0031) coasts of Korea and low value in the western coasts of Korea and Japan (only one haplotype), whereas C. ocellatus mostly shows a high diversity in the southern (H = 0.5059, π = 0.0015) and western (H = 0.5556, π = 0.0011) coasts of Korea and low diversity in the eastern Korea (H = 0.1947, π = 0.0008) and China (only one haplotype). In Jeju Island, both species exhibit relatively low genetic diversity, which has also been found in red macroalgae Pachymeniopsis spp. (Kim et al. 2014, Yang et al. 2021b). C. nipponicus was peculiar when compared to the other seaweeds studied, showing a slightly lower genetic diversity in the eastern coast than the southern coast (Kim et al. 2012). As for C. giganteus, with only two haplotypes the haplotype and nucleotide diversity were exceptionally low despite the high number of specimens (n = 139), similar to the results of Hu et al. (2015).
Haplotype networks are important to understand population structure in relation to distribution range (Cakil et al. 2021). We found that C. nipponicus (14 haplotypes) and C. ocellatus (11 haplotypes) share geographical distribution, although the most frequent haplotypes in each of the two species had different distributions. The major haplotype of C. ocellatus, CO1, had a more limited distribution in China and Japan than the major haplotype of C. nipponicus, CN1, although both species were found throughout Korea. In contrast, C. giganteus showed a unique haplotype distribution with one major haplotype, CG1 (97% of specimens), distributed only in a limited area in the southern Korea (PH, GJ, TY) and two sites in Japan (EN, NG). It is unclear whether the very low genetic diversity (H = 0.0563, π = 0.0002) in C. giganteus is due to a recent reduction in genetic diversity because of high gene flow or bottleneck effect, or due to a consistent historical lack of diversity (Yang et al. 2021b). Nevertheless, this low diversity raises concerns about its ability to adapt to climate change or other environmental alterations, which adds apparent difficulty to conservation efforts for natural seaweed resources (Liang et al. 2021, Yang et al. 2021b).
The estimated demographic history did not differ between C. nipponicus and C. ocellatus. The negative values of Tajima’s D and Fu’s FS may be reflect a recent selective sweep or population expansion (Tajima 1989, Fu 1997). Both processes may have affected Chondrus populations in the northwest Pacific Ocean during cold periods characterized by extreme reduction and fragmentation of suitable habitats (Hu et al. 2015). Each Chondrus species may have had opportunities to expand its population range after isolation by habitat fragmentation during the Last Glacial Maximum (Hu et al. 2015). Evidence of population expansion has often been found in marine species in the northern hemisphere including red algal species (Yang et al. 2021b, 2021c) including C. crispus (Hu et al. 2011, Provan et al. 2013). Such expansions have been suggested to result from the demographic effects of glaciations, particularly the most recent Last Glacial Maximum (Provan et al. 2013). The star-shaped haplotype networks of C. nipponicus and C. ocellatus also support a demographic scenario of sudden population expansion (Figs 2 & 3).
Based on our morphological findings, we conclude that the branching pattern of the thallus is not a valid diagnostic character for the three Chondrus species (Figs 2A, 3A & 4A). Thallus proliferations, traditionally used to identify C. nipponicus (Brodie et al. 1991), are also found in several individuals belonging to C. ocellatus in our observation. At the same time, some individuals of C. nipponicus lack these proliferations and have been erroneously identified as C. ocellatus. Additional observations of the cystocarp shape and position of tetrasporangial sori confirm the absence of diagnostic differences and limited variation in the three species, in contrast to previous findings (Mikami 1965, Brodie et al. 1991, 1993). Morphologically, the three species are indistinguishable from one another, although further morphological studies may reveal subtle distinguishing traits (Brodie et al. 1997). In accordance with our results, the extent of phenotypic variability within closely related species is unknown in many taxa, preventing accurate evaluation of diagnostic characteristics used for taxonomic identification (Furfaro et al. 2021).
The warm- and cold-temperate zones in the northwest Pacific are the center of the geographical distribution of the genus Chondrus (Hommersand et al. 1994). The geographical distributions of the three species (warm-temperate group) are influenced by warm currents (the Kuroshio Current and the Tsushima Current and its branches), some of which are also distributed in regions influenced by both warm and cold currents (the Kurile or the East Sakhalin currents). However, our results showed relatively shallow phylogeographic patterning for the three Chondrus species. Particularly, the haplotypes shared among the geographic regions suggest a high gene flow among regional groups, probably influenced by complex oceanic currents in the northwest Pacific. This gene flow was evidenced by the shared haplotypes CN1 of C. nipponicus, CO1 of C. ocellatus, and CG1 of C. giganteus, which were widely distributed in the collection regions (Figs 2C, 3C & 4C). This gene flow also exists in red algae from Korea such as two Pachymeniopsis species (Yang et al. 2021b) and two Grateloupia species (Yang et al. 2021c), which exhibited shared haplotypes and no significant population differentiation corresponding to geographic distribution. Since macroalgae have a relatively poor dispersal ability (Payo et al. 2013), the ocean currents around the Korean Peninsula are likely the main dispersal factor enhancing the gene flow in algal populations (Zhang et al. 2015, Yang et al. 2021b). Additionally, our study supported population differentiation between some regions. For example, the haplotype CN10 of C. nipponicus was distributed only in Korea, predominantly in the southwest coast (Fig. 2). For C. ocellatus, haplotype CO10 was found only in southern Japan, and the samples were genetically distinct from those collected in Korea, with no shared haplotypes. These patterns indicate that genetic diversification has been occurring with the recent increases in population size (Aris-Brosou and Excoffier 1996).
Understanding the level of genetic diversity within a species is essential to establish conservation strategies for the species (Hu et al. 2016). Although our collections covered only a limited part of the distribution range of Chondrus in the northwest Pacific, the present study provides insights into the phylogeographic pattern and may be helpful in the conservation of genetic diversity in this economically important seaweed. Our study also revealed similar phylogeographic patterns for C. nipponicus and C. ocellatus; they have a similar distribution range, moderate genetic diversity, and shallow phylogeographic structure with a star-shaped haplotype network, which are indicative of a recent population expansion. In contrast, C. giganteus has a limited distribution and extremely low genetic diversity, indicating potential vulnerability to environmental changes. Larger datasets will help to improve the phylogeographic structure in this region; therefore, we suggest continuous monitoring of the genetic diversity and phylogeographic pattern in the current populations for future conservation of these natural seaweed resources, including Chondrus.
SUPPLEMENTARY MATERIALS
Supplementary Table S1. Newly generated sequences information used to construct rbcL phylogenetic tree (https://www.e-algae.org).
Supplementary Table S2. Distribution of COI-5P haplotypes in populations of Chondrus nipponicus (https://www.e-algae.org).
Supplementary Table S3. Distribution of COI-5P haplotypes in populations of Chondrus ocellatus (https://www.e-algae.org).
Supplementary Table S4. Distribution of COI-5P haplotypes in populations of Chondrus giganteus (https://www.e-algae.org).