ABSTRACT
Cocconeis coreana and C. sijunghoensis are described as new based on micromorphological and molecular data. C. coreana is represented by five unialgal cultures from four different freshwater bodies, two from North Korea and three from South Korea. C. sijunghoensis is represented by two unialgal cultures from a brackish water body in North Korea. Except for one, all of the strains auxosporulated and showed an almost quadrupling of size in length and width. Morphologically, these species with their two different elliptical valves belong to the Cocconeis placentula group. The raphe valve has striae with uniseriate areolae continuing across a pronounced submarginal hyaline rim to the edge of the valve. The sternum valve has uniseriate dash-like areolae continuously from the valve face until the valve edge. Micromorphologically, these species possess two different open valvocopulae: only the raphe valvocopula has fimbriae; the sternum valvocopula has none. Based on p-distances of currently available DNA sequence data, i.e., rbcL and 18SV4, both species are pronouncedly different from the epitype strain of C. placentula, with C. coreana closest to the published molecular data of the strain UTEX FD23 named C. placentula from Iowa, USA, while C. sijunghoensis is closest but not the same as the published molecular data of strain D36_012, the epitype strain of C. placentula from Berlin, Germany. Based on scanning electron microscope observations, differentiating features are discussed concerning valvocopula fimbriae, central area, areolation of the sternum valve and on the raphe valve especially between the submarginal hyaline rim and edge.
INTRODUCTIONThe most ubiquitous and cosmopolitan freshwater diatom seems to be Cocconeis placentula Ehrenberg sensu lato; this taxon and its varieties are cited in almost every floristic study of fresh waters or water quality assessment from all over the world often occurring in large numbers and as pioneers on substrates (to cite just a few recent: Kobayasi et al. 2006, Gari and Corigliano 2007, Plenković-Moraj et al. 2008, Metzeltin et al. 2009, Leterme et al. 2010, Joh 2012, Hofmann et al. 2013, Potapova and Spaulding 2013, Abdel-Rahman and Sinada 2016, Ouyang et al. 2016, Taylor and Cocquyt 2016, Amutha and Muralidharan 2017, Lange-Bertalot et al. 2017, Mora et al. 2017, Toudjani et al. 2017, Pumas et al. 2018, Al-Handal and Al-Shaheen 2019, Chen et al. 2019, Heinrich et al. 2019, Jüttner et al. 2020).
In the Algal Flora of Korea, which is a compendium of all taxa from South Korea compiled until 2012, the author writes (Joh 2012, p. 108): “Cocconeis placentula and its neighboring diatoms are the most common in freshwater bodies and are to be cosmopolitan in lotic waters as rivers and streams, lentic ones of lakes, reservoirs and ponds in both freshwaters and estuarine waters.” These three neighboring diatoms are treated as varieties of Cocconeis placentula: C. placentula var. euglypta (Ehrenberg) Grunow, C. placentula var. lineata (Ehrenberg) Van Heurck, and C. placentula var. klinoraphis Geitler. Especially these four taxa are very difficult to differentiate by their morphology since features such as outline and striae counts overlap and true differentiating features are either not existing, or only visible in the scanning electron microscope (SEM).
The cells of Cocconeis placentula s.l. are monoraphid, having one thin valve with a raphe (termed raphe valve [RV]) and one thick valve with a sternum which in older literature used to be called pseudoraphe (termed sternum valve [SV]). Besides the raphe, uniseriate striae, and open valvocopulae which connect the valves, both valves are morphologically very different from each other. Until now, the morphology of the RV was understood to show no significant features to differentiate species. The striae of the RV consist of small circular areolae occluded by hymenes internally. The RV has a submarginal silicified hyaline rim towards the valve mantle; between this rim and the edge of the valve, areolae of different numbers and arrangements are placed; these details can only be seen in SEM. The SV have differentiating numbers of striae which are more or less densely punctated or dash-like, some more or less regularly separated from each other, sometimes producing longitudinal lines visible in the light microscope (LM). Only discernible in SEM, the punctae of the SV turn out to be deep areolae covered by hymenes externally, whereas the areolae are open to the inside and roundish in internal view. The raphe valvocopula has more or less simple fimbriae; there are no fimbriae at the sternum valvocopula (Jahn et al. 2009, Romero and Jahn 2013, plus further own observations; some of these features were already shown in the first electron microscopic study on Cocconeis by Holmes et al. 1982).
In diatom preparations from environmental samples when the two valves are separated, it is very difficult to differentiate these taxa. But unialgal cultures provide sufficient numbers of both valve types (SV and RV) of the same species to study the variability of features and evaluate their consistency. Thus, clonal cultures not only provide molecular data but also reveal some of the phenotypic plasticity of a single genotype. Molecular data are playing an important role in the discovery and delimitation of diatom species and are revealing a hidden diversity, cryptic taxa, resulting in the description of many new species (Mann et al. 2004, Alverson 2008, Kaczmarska et al. 2009, Poulićkova et al. 2010, Abarca et al. 2014, Jahn et al. 2017, 2019). There is also evidence that molecular sequence datasets yield not only significantly increased levels of taxonomic differentiation but also biogeographical resolution compared to analyses of morphological data alone (Wortley and Scotland 2006, Abarca et al. 2014, 2020, Skibbe et al. 2018).
In this paper we studied the morphological and molecular data of Cocconeis placentula s.l. strains which we had established from waters of the Korean Peninsula in the last 15 years.
The lectotypes of Ehrenberg’s Cocconeis species, C. placentula (Jahn et al. 2009), C. lineata, C. euglypta (Romero and Jahn 2013) and their epitype strains with their molecular and morphological data, serve as calibration reference for the new species from Korean waters. They are compared to further strain data available through the International Nucleotide Sequence Database Collaboration (INSDC) named C. placentula (Supplementary Table S1). Their micromorphologies and molecular data (18SV4, rbcL) plus morphometrics of seven strains and some information on auxospore formation have led to the description of two new species presented in this paper.
MATERIALS AND METHODSField collection and data of strainsEnvironmental brackish and freshwater samples were collected from North and South Korea between 2004–2014 (details in Supplementary Table S1). Seven monoclonal strains of Cocconeis specimens were established. Some data has been published before but under a different name (C. placentula or Cocconeis sp.): 18SV4 for five strains (Zimmermann et al. 2011); unfortunately, the molecular data of one of these strains (D26_016) is inaccurate and lack of reserve material prohibits resequencing but the morphological data is accurate. For calibration, molecular and morphological data of the epitype strains of C. placentula (Jahn et al. 2009), C. lineata, and C. euglypta (Romero and Jahn 2013) are used as reference. Six additional strains named Cocconeis placentula were retrieved from NCBI (e.g., Keck et al. 2016). All sequences available through the INSDC were compared in BLAST to infer similarity among sequences and to test for taxonomic consistency.
CultivationThe process of establishing our unialgal cultures was described in Zimmermann et al. (2014): water samples were observed under a SZH zoom stereo microscope (Olympus K.K., Tokyo, Japan) and single diatom cells were manually isolated by using capillary glass pipettes. Cells were then transferred to 5 cm diameter plastic Petri dishes containing culture medium (WC, Chu, and AlgaGrow). In order to separate cells and to remove attached particles, this treatment was repeated several times until microscopic inspection confirmed that pure cultures had been established. Cultures were kept at an ambient temperature of 18–22°C. Lumilux daylight 39 W T5 fluorescent tubes (OSRAM Licht AG, Munich, Germany ) from a distance of 1 m were used for illumination, except for strain D119_029 which was illuminated by 5000K LED panels.
Documentation of morphologyFor all newly established strains, the frustule preparation and morphological documentation were executed following Abarca et al. (2014). LM pictures were taken with a Axioscope or Axio Imager M2 plus AxioCam (Carl Zeiss AG, Oberkochen, Germany). Images of living cells were taken with a RED-1 digital motion camera on a Zeiss Photomicroscope III applying differential interference contrast. SEM pictures were taken with SEM 515 (Philips N.V., Amsterdam, Netherlands) (strain D26_016), Fei Quanta 200 (Thermo Fisher Scientific, Waltham, USA) (strains NK8_32) or SU810 FE-SEM (Hitachi, Tokyo, Japan) (all the other strains). Valve length, width, and density of striae and areolae of at least 20 valves per strain were measured using AxioVision Rel. 4.8 and ImageJ 1.47v software (Rasband 1997–2016) (see Table 1).
CurationVouchers and DNA of all strains are deposited in the collections at Botanischer Garten und Botanisches Museum Berlin, Freie Universität Berlin (B). DNA samples are stored in the Berlin DNA bank and are available via the Genome Biodiversity Network (GGBN)(Droege et al. 2014); nomenclatural acts are registered (Art. 42 of the International Code of Nomenclature for algae, fungi, and plants) (Turland et al. 2018) in PhycoBank (2017). Data are available through AlgaTerra (Jahn and Kusber 2005). Final DNA sequences were submitted to the ENA (http://www.ebi.ac.uk/ena/) using the software tool annonex2embl (Gruenstaeudl 2020).
DNA extraction, sequencing, and alignmentCultured material was transferred to 1.5 mL tubes. DNA was isolated using the NucleoSpin Plant II Mini Kit (Macherey and Nagel, Düren, Germany) following the manufacturer’s instructions. DNA fragment size and concentrations were measured via gel electrophoresis (1.5% agarose gel) and Nanodrop (PeqLab Biotechnology, Erlangen, Germany), respectively. The DNA samples were stored at −20°C for future use and finally deposited in the Berlin collection of the DNA bank network (Droege et al. 2014). Amplification of the gene rbcL was conducted via polymerase chain reaction (PCR) following Abarca et al. (2014). The V4 region of the 18S small subunit rRNA gene (18SV4) was amplified following Zimmermann et al. (2011). PCR products were visualized in a 1.5% agarose gel and cleaned with MSB SpinPCRapace (Invitek LLC, Berlin, Germany) following standard procedures. DNA concentrations were measured using Nanodrop (PeqLab Biotechnology) and samples were normalized to a total DNA content >100 ng μL−1 for sequencing. Sanger sequencing was conducted by Starseq (GENterprise, Mainz, Germany). The rbcL gene was sequenced according to Abarca et al. (2014) and 18SV4 was sequenced following Zimmermann et al. (2011). In both cases the same primers were used for amplification and sequencing. The editing, as well as the quality control of the pherograms for the new sequences, were done in Phyde (Müller et al. 2010). The evaluated sequences were aligned using MUSCLE (Edgar 2010), as implemented in MEGA X (Kumar et al. 2018). The alignments also included sequences from INSDC (Supplementary Table S1). Uncorrected p-distances were calculated for each marker individually using the default settings implemented in MEGA X (Kumar et al. 2018). Those parts of the alignment where data are missing were excluded from the analysis as missing data affect the comparability when calculating uncorrected p-distances from sequences with different informative content (Tables 2 & 3).
RESULTSVoucher and geographic data of our studied strains are summarized in Supplementary Table S1; morphometric data in Table 1; data of p-distances for rbcL and 18SV4 markers in Tables 2 & 3. Pictures of alive cells including auxospore formation of both species are shown in Fig. 1, of valves in LM in Figs 2–6 & 12, and of valve details in SEM in Figs 7–11, 13 & 14. The molecular and morphometric results from our epitype strains published earlier have been included for comparison: C. placentula sensu stricto strain D36_012 (Jahn et al. 2009) as well as C. euglypta strain WiCoc2a and C. lineata strain D17_011 (for C. lineata strain D17_011 the striae counts per 10 μm are here corrected to 22–28 and 24.5 average) (Romero and Jahn 2013). Comparable data from INSDC for strains named C. placentula and its varieties have also been included.
Strains D119_029, K13rj7_9, D26_016, KCoc12_1, and KCoc12_2 (Figs 1A–G & 2–11)Morphologically, four strains (D119_029, K13rj7_9, D26_016, and KCoc12_1), produced auxospores which means that the morphometric measures represent the entire size range from the smallest to the largest with only a few intermediate sized valves present; striae counts deviate conspicuously between small and large valves with lower counts on the large valves. The summarized morphometric data for the five strains D119_029 (Figs 2, 7A, B, D, E, 8, 9B–G, 10A, B & 11), K13rj7_9 (Figs 1A–D, 3, 7C, 9A, 10D & E), D26_016 (Figs 4, 10C & F), KCoc12_1 (Figs 1E–G & 5) and KCoc12_2 (Fig. 6) are as follows: length (AA) 15.4–59.3 μm, width (TA) 9.9–43.1 μm; with a broad length to width ratio i.e., 1.32–2.7. The number of striae is similar but the number of areolae is different on each valve, 18–26 striae per 10 μm on the RV; 17–26 striae per 10 μm on the SV. The striae on the RV are punctate and regularly spaced with 10–24 areolae in 10 μm. The striae on the SV are very short dash-like with 8–16 areolae per 10 μm (for morphometric details of each strain) (Table 1).
Molecularly, there are some discrepancies of two strains to the other three. The data for D26_016 (18SV4) has been published before (Zimmermann et al. 2011) but is incomplete, rbcL data is not available because there is no more DNA material; therefore, this strain will be excluded from the molecular discussion. The other four strains have quality data for the markers 18SV4 and rbcL. Three show the exact same sequence data in the respective markers; strain KCoc12_2 has only one base-pair difference in rbcL (0.2%) but 1.1% in 18SV4. Comparing these data with the INSDC data of the strains named C. placentula for 18SV4 and rbcL (see Supplementary Table S1; our rbcL newly submitted), the strain UTEX FD23 named Cocconeis placentula from Iowa, USA, is the closest with 2.6–2.8% p-distances for rbcL (Table 2) and 6.1% for 18SV4 (Table 3). The p-distances to the other Cocconeis strains are higher, ranging between 4.0–8.3% for rbcL (Table 2) and between 6.9–10.1% for 18SV4 (Table 3).
Cocconeis coreana R. Jahn & B. M. Suh sp. novHolotypeSlide B 40 0041501a, Botanic Garden and Botanical Museum Berlin, from the strain D119_029. Fig. 2F (SV; Fig. 2B RV) represents the holotype. SEM-stub deposited as B 40 0041501b. For molecular material and data see “Materials and Methods” and Supplementary Table S1.
Type localityDPR Korea, Myohyangsan, Hyangsan waterfalls, Sep 17, 2014, collected by Byung Moon Suh & Regine Jahn.
EtymologyWe are referring to the entire Korean Peninsula since this taxon was found in clean freshwaters in North and South Korea.
Description by LM (type strain only, Fig. 2)Valve outline is generally slender elliptical (Fig. 2A) but sometimes round elliptical (Fig. 2B), apices are obtusely rounded (length [AA]: 25.5–59.3 μm; width [TA]: 14.1–36.6 μm [n = 34]). The number of striae on both valves are similar but the individual punctae are different. The RV shows punctate striae with 18–24 rows per 10 μm and 15–24 areolae in 10 μm. The SV shows short dash-like striae with 18–24 rows per 10 μm and 9–12 areolae per 10 μm. The striae on both valves are parallel at the center and radiate near the apices; the striae of the SV seem to be in bundles of 2–5 striae (Fig. 2E–H). The RV shows a pronounced round central area (Fig. 2A–D). Only the RV has a rim of several rows of areolae at the valve mantle as a continuation of the striae separated by a submarginal hyaline rim (for details see SEM). In addition, the RV has a conspicuous valvocopula with irregular spaced clamp-like fimbriae (for details see SEM). Raphe is filiform and straight, axial area very narrow; sternum axial area very narrow. Concerning alive cells, in one strain we documented a single c-shaped plastid with serrated edges and a pyrenoid (Fig. 1A–C) and in two strains we observed auxospore formation (Fig. 1D–G) (not for the type strain).
Description by SEM (Figs 7–11)RVThe valve face is flat with a concave mantle separated by a pronounced hyaline rim, well visible externally (Fig. 7A–C) and internally (Fig. 8A & C). Striae are made up of round to slightly elliptical uniseriate areolae which are small (about 0.25 μm) and regularly spaced, increasing in size from the axial area towards the valve margin and the apices (Fig. 7E), forming a pronounced central area of about 6 rows of areolae length (AA) and 3 areolae wide on each side of the raphe ends (TA) (Fig. 7A–C). Internally, the areolae are occluded by round lattice-structured hymenes with short slits around the margin and tiny perforations in the middle (Figs 7D, 8B & D). On the mantle, the uniseriate valve face striae continue as uniseriate rows of two to four, generally three areolae (Fig. 7A–C); at each pole there is a gap of these mantle areolae (Fig. 7E). Externally, the raphe is straight, filiform, proximal raphe endings are slightly drop-like expanded (Fig. 7A–C). Internally, the proximal raphe fissures are bent to opposite sides (Fig. 8A & C); distal raphe endings are straight and end in weakly elevated helictoglossae on the hyaline rim (Fig. 8A & C). Valvocopula is open; long, slender and heavily silicified clamp-like fimbriae (without papillae and furrows) seem to be attached to the hyaline rim and are irregularly spaced at every 4th to 7th stria, or 3 to 5 clamps per 10 μm (Figs 8A & 10F).
SVThe valves are thicker than the RV (compare Figs 7D & 9G). The valve face is flat to slightly convex with a short mantle (Fig. 9A). Striae are made up of dash-like regular uniseriate areolae, undifferentiated unto the valve edge (Fig. 10A & D); internally they are rounded (Fig. 9D & F) whereas externally they are slit-like of different openness (Fig. 10B & E). Areolae are occluded by elongated lattice-structured hymenes of about 0.7 μm length by 0.13 μm width with marginal slits and tiny holes in the center; the hymenes lie towards the outer side but not completely externally (Figs 9E, G & 10B). The sternum is very thin and sometimes shows the mark of a ghost raphe (Fig. 10A). The valvocopula is open with no visible fimbriae (Figs 9B & 10C; the fimbriae and valvocopula seen in Fig. 9C are from the RV).
We also observed auxospore formation in SEM (Fig. 11) with remnants of incunabula (Fig. 11A & B) but apparently no perizonium.
Comparison to related taxaWith its largest length of almost 60 μm and width of up to a 43 μm C. coreana is generally bigger than C. placentula var. placentula (sec. Jahn et al. 2009, figs 28–43). Nevertheless, their length to width ratios overlap, with C. coreana having a broader ratio (see morphometric values in Table 1). The RV of C. coreana looks like a replica of C. placentula with generally similar numbers of striae and areolae, becoming larger towards the pronounced hyaline rim; also the numbers of areolae of the mantle below the hyaline rim are similar with about 3 (2–4 for C. coreana and 3–4 for C. placentula) per row but there is a gap at both poles in C. coreana (Fig. 7A–C) in contrast to continuing areolae in C. placentula (Jahn et al. 2009, figs 37 & 39). Further, in C. coreana the numbers and spacing of the longer clamp-like fimbriae of the raphe valvocopula are similar but there seem to be no small fimbriae in C. coreana (Figs 8A & 9C) in contrast to C. placentula (Jahn et al. 2009, fig. 42). The size of the central area around the drop-like proximal raphe ends is pronouncedly bigger in C. coreana (Fig. 7A–C) than in C. placentula (Jahn et al. 2009, figs 37 & 39). As expected for the genus Cocconeis, the biggest differences are found in the SV concerning numbers of the dash-like striae, size of the areolae and size of the sternum: C. coreana has a narrow sternum and closely spaced areolae of the size 0.6 μm by 0.2 μm externally (Fig. 10C); C. placentula has a broad sternum and widely spaced long and very thin areolae of the size of up to 1.5 μm by 0.05 μm externally (Jahn et al. 2009, figs 40 & 41).
Comparing C. coreana to C. lineata (sec. Romero and Jahn 2013, figs 2–8) is problematic since the only available data is from one strain without auxospores. C. lineata has a higher length to width ratio (see morphometric values in Table 1) which means that C. lineata is smaller and slenderer in comparison to C. coreana. The mantle of the RV of C. lineata shows 2–3 rows of areolae with a gap at the poles as C. coreana. The length to width ratios and the numbers and density of the SV areolae are similar in C. coreana and C. euglypta (sec. Romero and Jahn 2013, figs 9–18) (see morphometric values in Table 1). In C. euglypta the hyaline rim is barely visible externally and the raphe mantle areolae rows are only 2 in C. euglypta.
Strains NK8_32a and NK8_32b (Figs 1H–O & 12–14)Morphologically, both strains produced auxospores, representing the smallest and the largest size ranges with only a few intermediate sized valves observed. Striae counts deviate conspicuously between small and large valves with lower counts on the large valves. The summarized morphometric data for the two strains NK8_32a (no Figs) and NK8_32b (Figs 1H–O & 12–14) are as follows: length (AA) 11.5–35.6 μm, width (TA) 7.6–27.4 μm; with a narrow length to width ratio, i.e., 1.13–1.6 (for morphometric details of each strain, see Table 1).
Molecularly, the two strains show the same sequence data for the marker 18SV4 as well as for rbcL; the 18SV4 data has been published before (Zimmermann et al. 2011). Comparing these data with the available data of C. placentula strains for 18SV4 and rbcL (see Supplementary Table S1; rbcL newly submitted), our epitype strain D36_012 Cocconeis placentula from Berlin, Germany, is the closest with only a p-distance of 1.9% for rbcL (Table 2) and of 2.6% for 18SV4 (Table 3); for 18SV4, it also has a distance of 2.6% to TCC501 and TCC518 (both named Cocconeis placentula in INSDC). All the other strains named Cocconeis placentula have higher p-distances between 2.2–8.9% for rbcL (Table 2) and between 4.8–8.8% for 18SV4 (Table 3).
Cocconeis sijunghoensis R. Jahn & B. M. Suh sp. novHolotypeSlide B 40 0040811a, Botanic Garden and Botanical Museum Berlin, from the strain NK8_32b. Fig. 12F (SV; Fig. 12A RV) represents the holotype. SEM-stub deposited as B 40 0040811b. For molecular material and data see Materials and Methods and Supplementary Table S1.
Type localityDPR Korea, Lagoon Si Jung Ho, collected by Byung Moon Suh & Regine Jahn on Apr 24, 2011.
EtymologyWe are referring to the beautiful Lagoon Si Jung Ho which is situated very close to the East Sea in North Korea.
Description by LM (Figs 1H–O & 12)Valve outline is round elliptical (Fig. 12G) but mostly just elliptical and somewhat lopsided (Fig. 12F), apices are round or obtusely rounded (length [AA]: 12.9–35.6 μm; width [TA]: 8.9–27.4 μm [n = 60]). The number of striae is similar but the number of punctae in the stria are different on each valve. The RV has punctate striae with 22–28 rows per 10 μm and 18–25 areolae in 10 μm which are barely resolvable in LM. The SV has very short dash-like striae with 21–30 rows per 10 μm and 10–19 areolae per 10 μm. The striae on both valves are slightly radiate at the center and strongly radiate near the apices; the striae of the SV seem to be in fascia of up to 7 striae; the RV shows a very small round central area (Fig. 12A). Only the RV has a rim of several areolae at the valve mantle as a continuation of the striae separated by a hyaline ring (for details see SEM). In addition, the RV has a conspicuous valvocopula with finger-like fimbriae irregularly spaced (for details see SEM). Raphe filiform and straight, axial area very narrow; sternum axial area pronounced and narrow. We observed auxospore formation in the alive strain (Fig. 1H–O) and documented one chloroplast in u-form.
Description by SEM (Figs 13 & 14)RVThe valve face is flat with a weakly concave mantle separated by a hyaline rim (Fig. 13A, B, D & E), visible externally and internally. Striae are made up of round to slightly elliptical uniseriate areolae which are small and regularly spaced, increasing slightly in size from the axial area towards the valve margin and the apices (Fig. 13A), forming a small lopsided central area of about 2 rows of areolae length (AA) and 1 areola wide on each side of the raphe ends (TA). Internally, the areolae are occluded by round lattice-structured hymenes of about 0.25 μm (Fig. 13F). On the mantle, the uniseriate valve face striae continue as uniseriate rows of one to three areolae (Fig. 13A & C); at the poles there are no real gaps of these mantle areolae (Fig. 13A). Externally, the raphe is straight, filiform, proximal raphe endings are very slightly expanded (Fig. 13A). Internally, the raphe lies in a slightly elevated silicified rim and the proximal raphe fissures are slightly bent to opposite sides (Fig. 13D & E); distal raphe endings are straight and lie in weakly elevated helictoglossae just short of the hyaline rim (Fig. 13D & E). Valvocopula is open; irregular spaced finger-like fimbriae can be noted (Fig. 13C).
SVThe valve face is flat to slightly convex with a short mantle (Fig. 14A). Striae are made up of short dash-like regular uniseriate areolae (Fig. 14A); internally they are roundish (Fig. 14C) whereas externally they are slit-like (Fig. 14B). Areolae are occluded by elongated lattice-structured hymenes of 0.55 μm length by 0.1 μm width with marginal slits and tiny holes; the hymenes lie towards the outer side (Fig. 14B). The sternum is pronounced (Fig. 14C). The valvocopula is open and has no visible fimbriae (Fig. 14C; the fimbriae seen on the left side in Fig. 14C are from the RV).
Comparison to related taxa
C. sijunghoensis and C. placentula var. placentula (sec. Jahn et al. 2009, figs 28–43) show an almost complete overlap in size with C. sijunghoensis being pronouncedly wider which is reflected in lower length to width ratios. The numbers and density of the SV areolae are much higher in C. sijunghoensis (see morphometric values in Table 1). In addition, on the RV the raphe mantle areolae are only 1–3 rows in C. sijunghoensis in contrast to 3–4 rows for C. placentula and both are continuous at the poles (without a gap). Both species have irregular spaced finger-like fimbriae on the raphe valvocopula but there seem to be no short fimbriae in C. sijunghoensis.
For C. lineata (sec. Romero and Jahn 2013, figs 2–8) length to width ratios do not overlap with C. sijunghoensis which means that C. sijunghoensis is much rounder and broader than C. lineata (see morphometric values in Table 1).
C. sijunghoensis is smaller and wider than C. euglypta (sec. Romero and Jahn 2013, figs 9–18) with length to width ratios overlapping only little (see morphometric values in Table 1). In C. euglypta the hyaline rim of the RV is thin and barely visible externally, with 2–3 rows of areolae at the raphe mantle.
DISCUSSIONThe Algal Flora of Korea (Joh 2012) lists C. placentula plus three neighboring taxa as its varieties: var. euglypta, var. lineata, and var. klinoraphis. Some of the many LM pictures of these taxa with their very different outlines and striation could represent both new species described here, C. coreana and C. sijunghoensis. But their true differentiating features can only be discerned by SEM. Nevertheless, two SEM pictures (Joh 2012, fig. 112A & B) show size, outline, expanded central area, hyaline rim with 2–3 areolae plus gap at the poles, which are features that could represent C. coreana. Concerning C. sijunghoensis, most of its cells are somewhat lopsided resembling C. placentula var. klinoraphis (Fig. 12F) but their striae numbers on the SV are higher than the pooled data for C. placentula including its varieties sec. Joh (2012). We also noticed this unregular match, –inclination– of raphe to sternum axial area, in a few valves of C. coreana (Fig. 3B).
Comparing C. coreana and C. sijunghoensis to other morphologically similar taxa is difficult because, on the one hand, our study provides no data on the morphological variability of the newly described species in the environment before undergoing cultivation. On the other hand, no material derived from clonal culturing is available in current studies on freshwater Cocconeis taxa (e.g., Stancheva 2019, Wetzel et al. 2019, Costa et al. 2020). For this reason, molecular data are missing and morphological changes during the life cycle have not been documented. The presumed single species populations in environmental samples do not give us the certainty that we are using morphologically stable features for the differentiation of species. For example, in the paper by Costa et al. (2020) the high quality SEM figs 25–28 of C. amerieuglypta L. F. Costa, C. E. Wetzel & Ector are, in our understanding, more than one species (but which valve represents C. amerieuglypta?) indicated by the differentiating feature numbers of areolae plus gap below the hyaline rim. In addition, further important features such as valvocopula structure (open or closed, with or without fimbriae), and occlusion structures (hymenes or vela) of the areolae (Holmes et al. 1982) are missing as in many other publications including some of our own (i.e., Romero and Jahn 2013). A recent paper lectotypifies an old taxon named C. rouxii Héribaud & Brun with an apparently restricted occurrence in the Massif Centrale, France, which also used to be regarded as a variety of C. placentula; the SEM pictures with details on valvocopulae, fimbriae, and hymenes of RV and SV demonstrate that detailed SEM studies can support the differentiation of taxa in this group (Wetzel et al. 2019). Another recent paper describes a new species from California, C. cascadensis Stancheva, and detailed SEM findings show that this differentiation seems justified (Stancheva 2019).
Neither many Cocconeis taxa have been cultivated nor the related molecular data have been published yet. Besides our own studies on four Ehrenberg taxa (Jahn et al. 2009, Romero and Jahn 2013), epitypified with strains from the River Spree in Berlin (C. placentula, C. pediculus), from a freshwater creek on the Faroe Islands (C. lineata), from the coast of the brackish Baltic Sea at Wismar (C. euglypta, Cocconeis sp.), molecular data of five strains are available (see Supplementary Table S1) which are one strain from an inland water in Iowa, USA (Ruck and Theriot 2011), and four from inland waters of the Island Mayotte, Indian Ocean, France (Kermarrec et al. 2013, Keck et al. 2016). According to these presently available rbcL and 18SV4 sequence data, C. coreana and C. sijunghoensis do not coincide with any published Cocconeis taxa. The p-distances between both species are 4.6–4.7% for rbcL and as high as 8.8% for 18SV4, respectively (for details see Tables 2 & 3). All the other similar looking taxa have at least p-distances of 1.9% for rbcL and 2.6% for 18SV4, respectively. The significantly different p-distances indicate that the two Korean species needed to be described as new, which is confirmed by micromorphological differences observed under the SEM. Even though there are some base-pair differences of KCoc12_2 (p-distances of 0.2% for rbcL and 1.1% for 18SV4) to the other three strains of C. coreana, we treated all as the same species since no morphological differences were obvious.
The here presented SEM and molecular data are further evidence that the C. placentula complex contains many different species even though in LM they all look similar with differences only in outline and striation of the SV. Concerning their typical, mostly elliptical outline, the size series of each strain from the smallest to the first valves after auxosporulation show an almost quadrupling of the valve size as well as decisive changes of valve outline, with the largest being most different and the smallest most similar to all the other species from the C. placentula complex. The typical striation of the SV is produced by the striae number and areolae density but its counting also depends on the focus either on the interior or the exterior of the heavily silicified valve. Even the RV which is generally understood of being the same in all species, shows decisive differences especially concerning the numbers of areolae per striae, the rows of areolae between the more or less pronounced submarginal hyaline rim and the edge of the valve, and the valvocopula.
Six of our seven strains auxosporulated so that we were able to document small and large valves (Figs 2–6 & 12) plus initial valves and auxospores plus incunabula but apparently no perizonium (Figs 1 & 11). Since our priority was on cultivation for molecular and micromorphological data, we did not conduct cytological studies and nuclear staining. Therefore, we cannot compare our incomplete auxospore data to the published observations of Geitler (1973) or Mizuno (1998) but used Kaczmarska et al. (2013) for orientation. Nevertheless, our live and SEM observations provide valuable information as reference for future studies.
Concerning their distribution, we found C. coreana five times in four different water bodies in North and South Korea in four different years (2004, 2012, 2013, 2014) which shows that this species must be widely distributed on the Korean Peninsula. This is in contrast to the new species C. sijunghoensis which we found only in one year in one water body in North Korea and for which we could find no real match in the presented data and taxa of South Korea (Joh 2012).
As we have demonstrated in previous papers on taxa in the genera Planothidium, where we described four new species for Korea (Jahn et al. 2017), Gomphonema, where we described one new species for Korea (Abarca et al. 2014, 2020), Gomphonella (Jahn et al. 2019), and in this paper, many more micromorphological differentiating features can be established, which are supported by molecular data, when strains of target species are cultivated. With more Cocconeis placentula s.l. strains from a broader set of types of waters from all over the world, we will be able to better understand the biogeography and phylogeny of these taxa.
ACKNOWLEDGEMENTSThe authors are grateful to Dr. Suh Byung Moon for his contribution to the description of these new species and for his longtime support in collecting diatoms from Korea. They also thank Jana Bansemer for work in the molecular lab, Juliane Bettig and Monika Lüchow at the SEM, and Michael Rodewald for help with the plates. The authors acknowledge funding by the Friends of the BGBM for cultivation support, the Deutsche Forschungsgemeinschaft for SEM [INST130/839-1FUGG], for Names Registration [JA874/8-1], and by the Bundesministerium für Bildung und Forschung [01LI1501E] for GBOL2 Diatoms, and by the Swedish Research Council for Sustainable Development (FORMAS) for cooperation.
SUPPLEMENTARY MATERIALSSupplementary Table S1List of materials: strain number, voucher code, taxon name, DNA bank and INSDC accession numbers, and locality (https://www.e-algae.org). Fig. 1
Cocconeis coreana and Cocconeis sijunghoensis, living cells, differential interference contrast microscope. (A–G) C. coreana. (A–C) Three focal planes of a small cell before auxosporulation. C-shaped plastid with serrated edges and single pyrenoid (Py). (D) Side view of early monoparental auxospore with attached sternum valve (SV) on top and raphe valve (RV) below. Inside the auxospore the initial cell’s dome-shaped SV is formed (arrowheads). (E–G) Three focal planes of fully developed auxospore surrounded by incunabula (arrows). (H–O) C. sijunghoensis. (H–J) Three focal planes of small cells before auxosporulation. C-shaped plastid (Pl) and nucleus (Nu) visible. (K) Side view of developed auxospore containing initial cell and first post-initial cell. Initial cell with dome-shaped SV (SV1), SV of post-initial cell (SV2) only slightly convex. RVs of initial and post-initial cells (RV1 + RV2) are face to face. (L–N) Development of mono-parental auxospore. (O) Post-initial cell released from auxospore. (A–D), strain K13rj7_9; (E–G), strain KCoc12_1; (H–O), strain NK8_32. Scale bars represent: A–J & L–O, 10 μm. 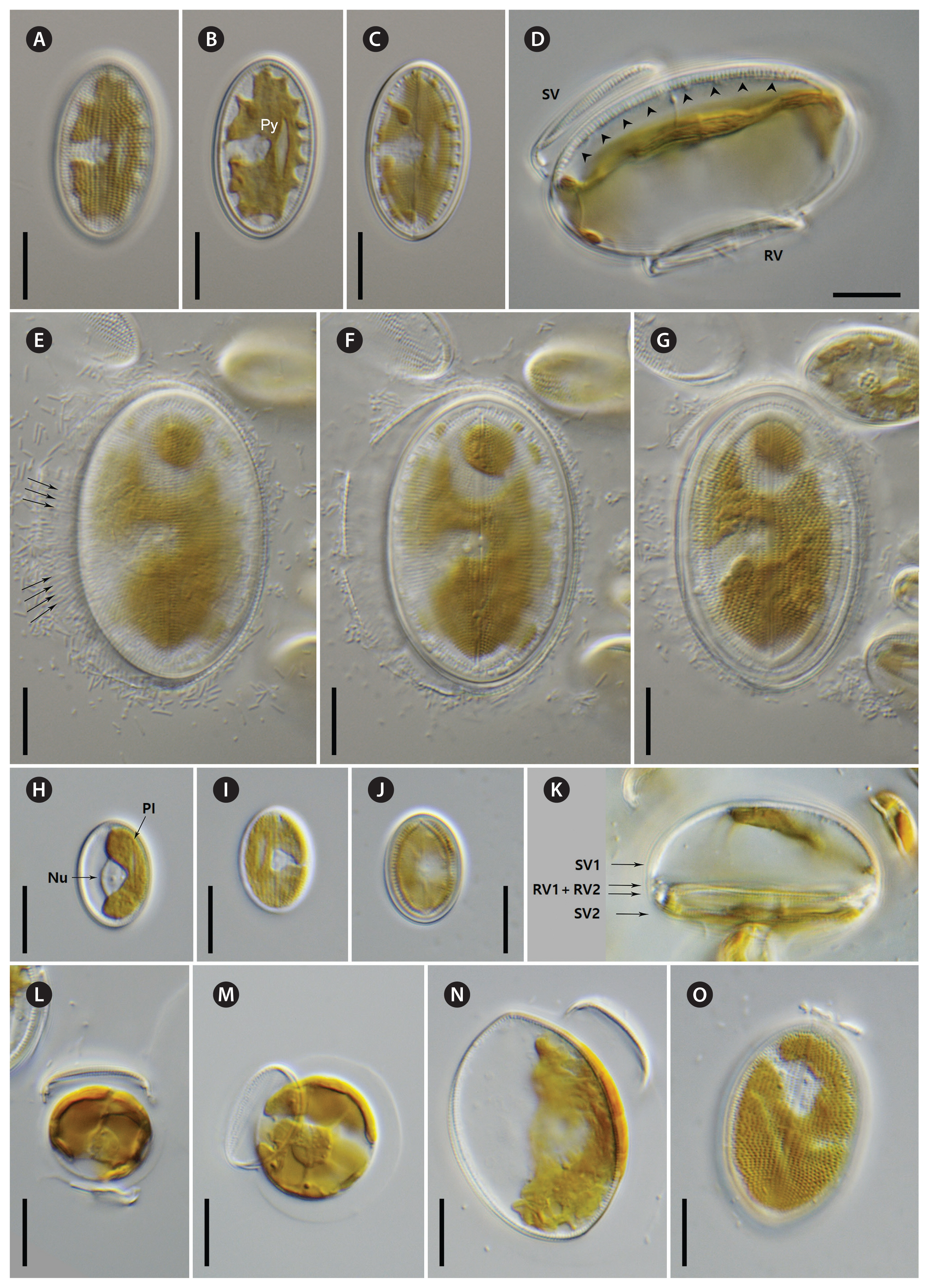
Fig. 2
Cocconeis coreana, strain D119_029 (type strain), light microscopy. (A–D) Raphe valve (RV). (B & C) Note the valvocopula with irregular fimbriae. (E–H) Sternum valve (SV). (F) (SV; Fig. B RV) represents the holotype. Scale bar represents: A–H, 10 μm. 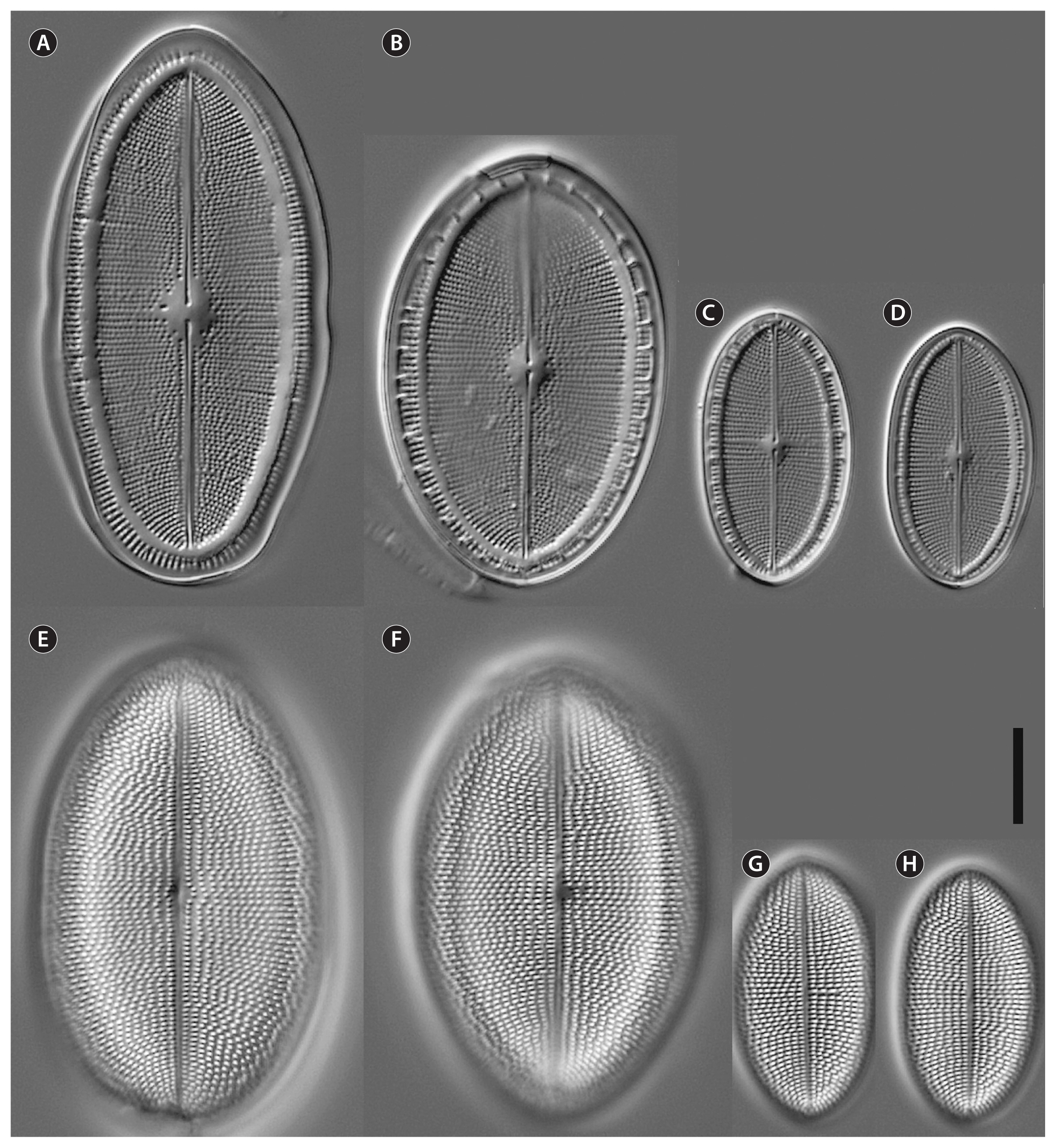
Fig. 3
Cocconeis coreana, strain K13rj7_9, light microscope. (A–C & G–I) Raphe valve. (B) Note the lopsided raphe in contrast to the sternum behind. (D–F & J–L) Sternum valve. Scale bar represents: A–L, 10 μm. 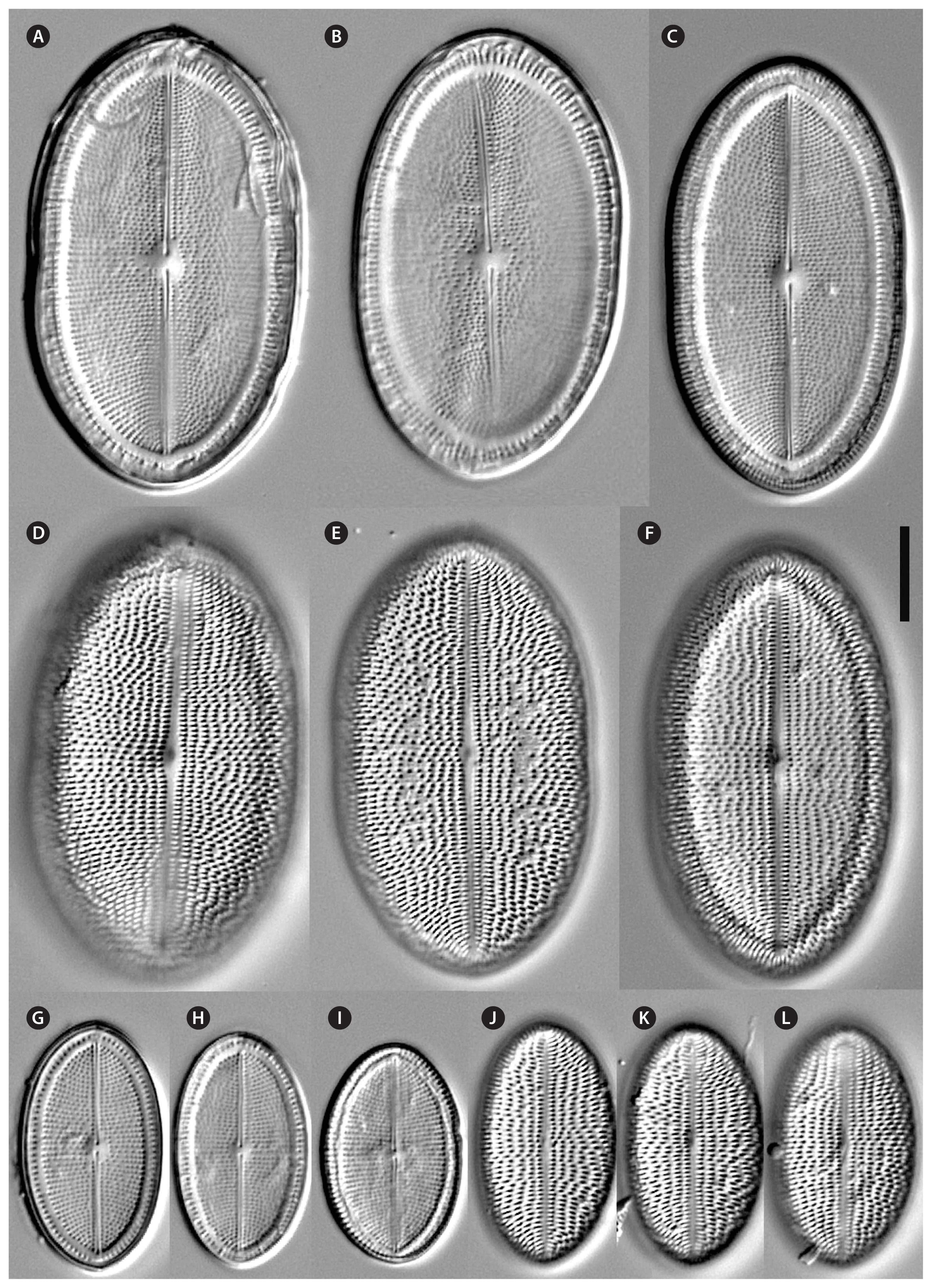
Fig. 4
Cocconeis coreana, strain D26_016, light microscope. (A–D) Raphe valve. (E–H) Sternum valve. Scale bar represents: A–H, 10 μm. 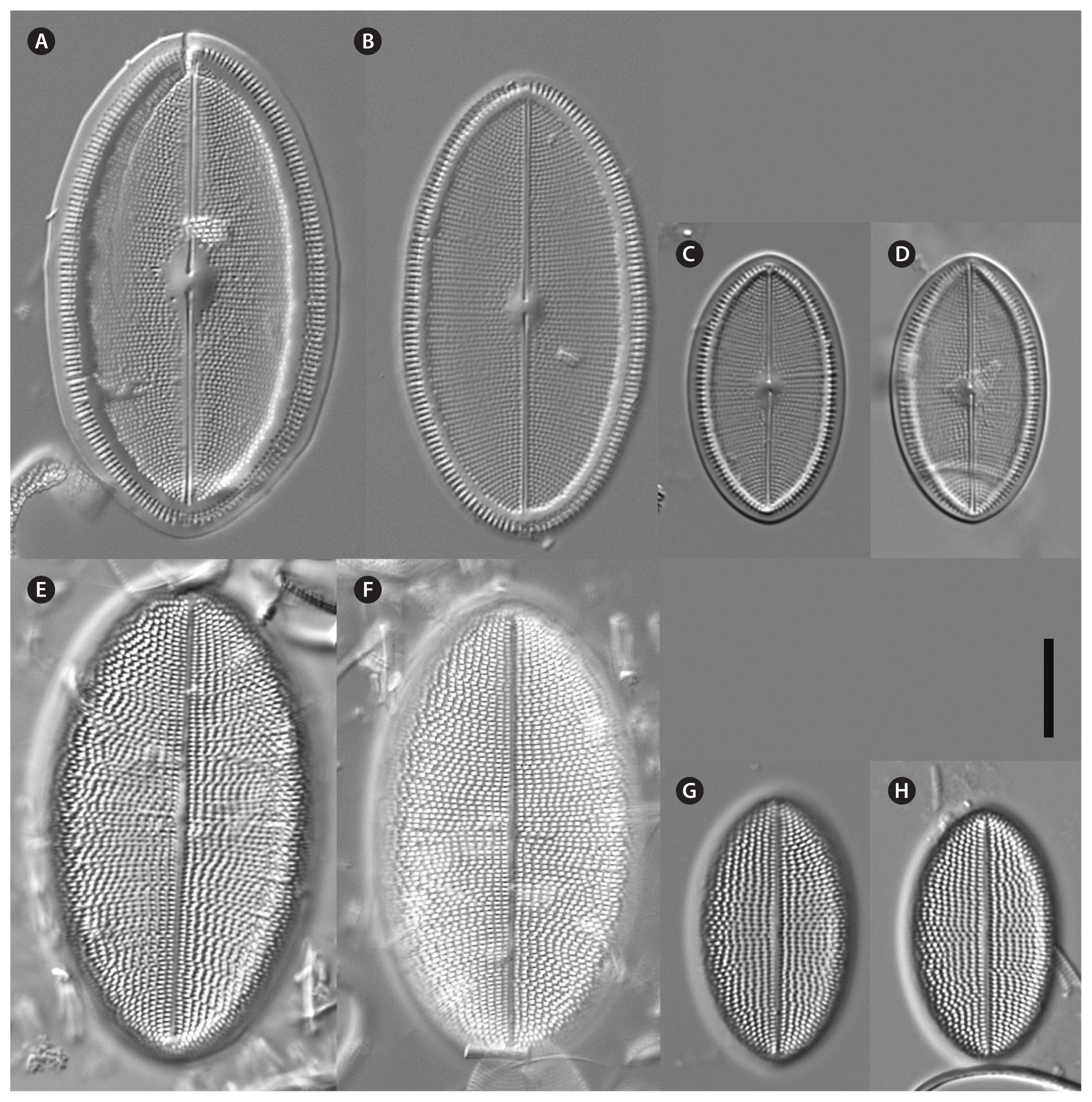
Fig. 5
Cocconeis coreana, strain KCoc12_1, light microscope. (A–D) Raphe valve. (E–H) Sternum valve. Scale bar represents: A–H, 10 μm. 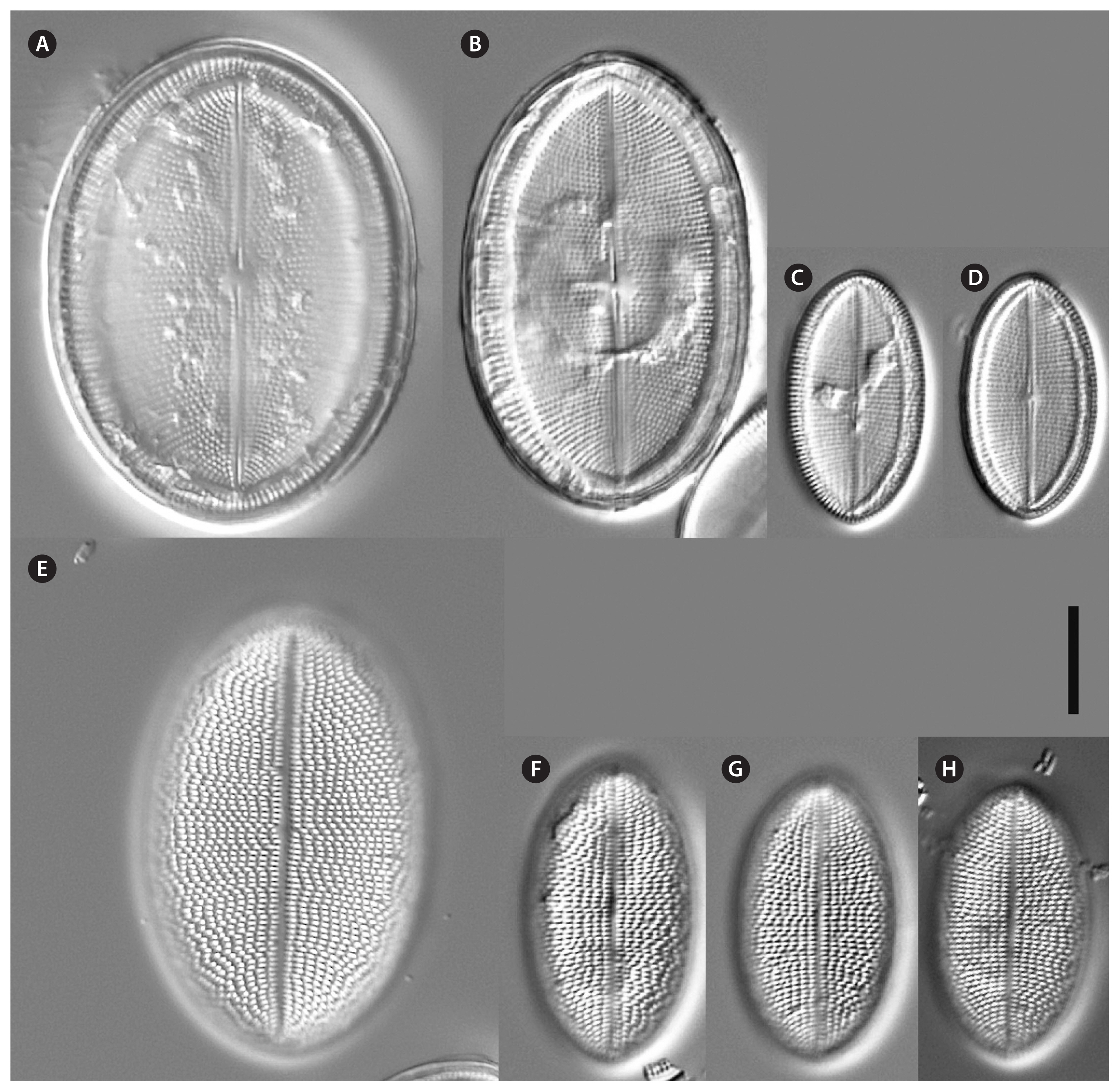
Fig. 6
Cocconeis coreana, strain KCoc12_2, light microscope. (A–F) Raphe valve. (G–L) Sternum valve. Scale bar represents: A–L, 10 μm. 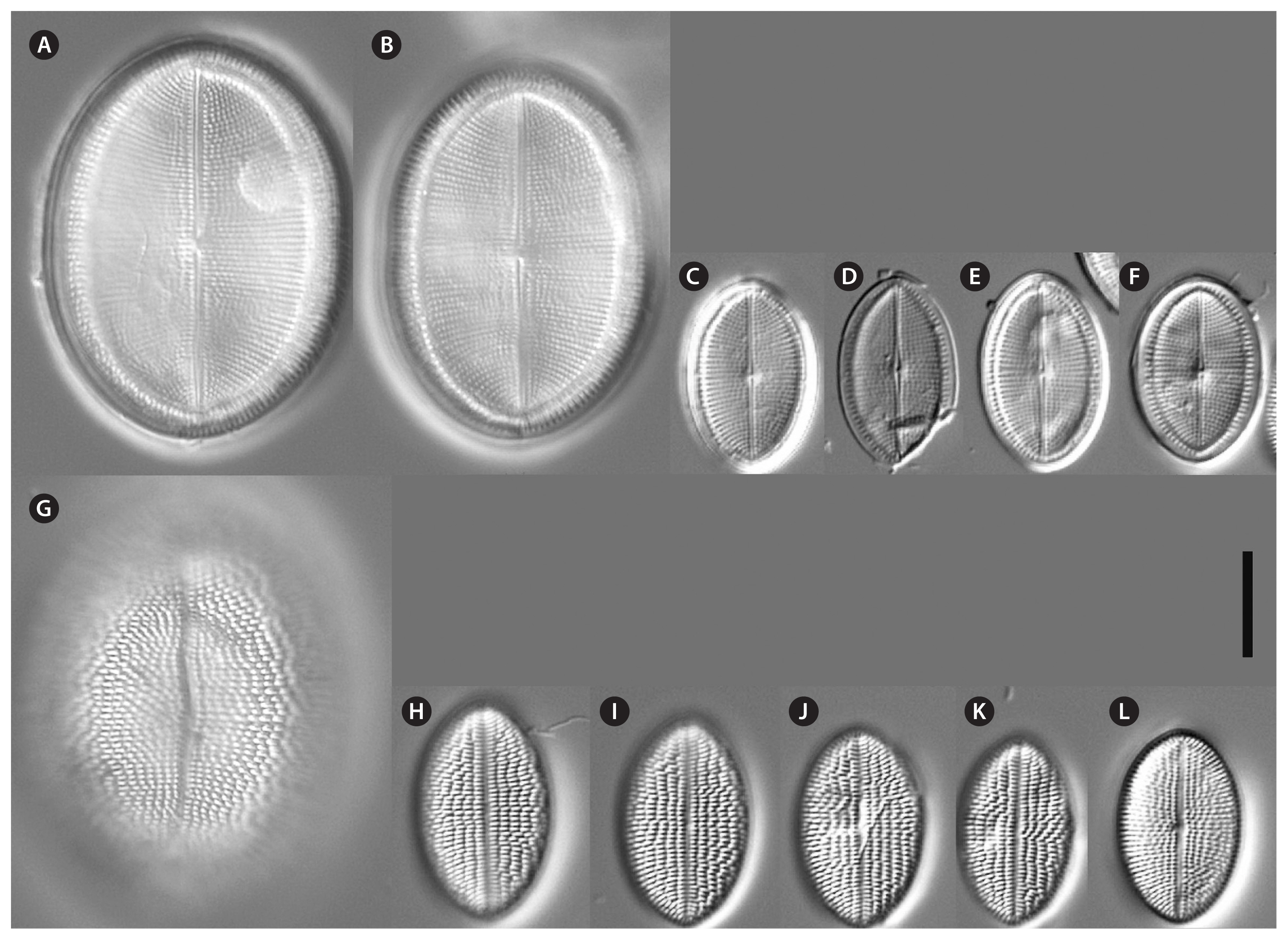
Fig. 7
Cocconeis coreana, scanning electron microscope. (A–E) Raphe valve external views of different sizes. (A–C) Uniseriate striae continue as round areolae across the hyaline rim unto the mantle; note the enlarged round central area and the slim axial area. (D) The areolae are open externally and covered with a (here broken) hymen internally. (E) Note the numbers of areo lae in the striae between rim and edge and the gap of areolae at the apices. (A, B, D & E), strain D119_029; (C), strain K13rj7_9. Scale bars represent: A–C & E, 10 μm; D, 1 μm. 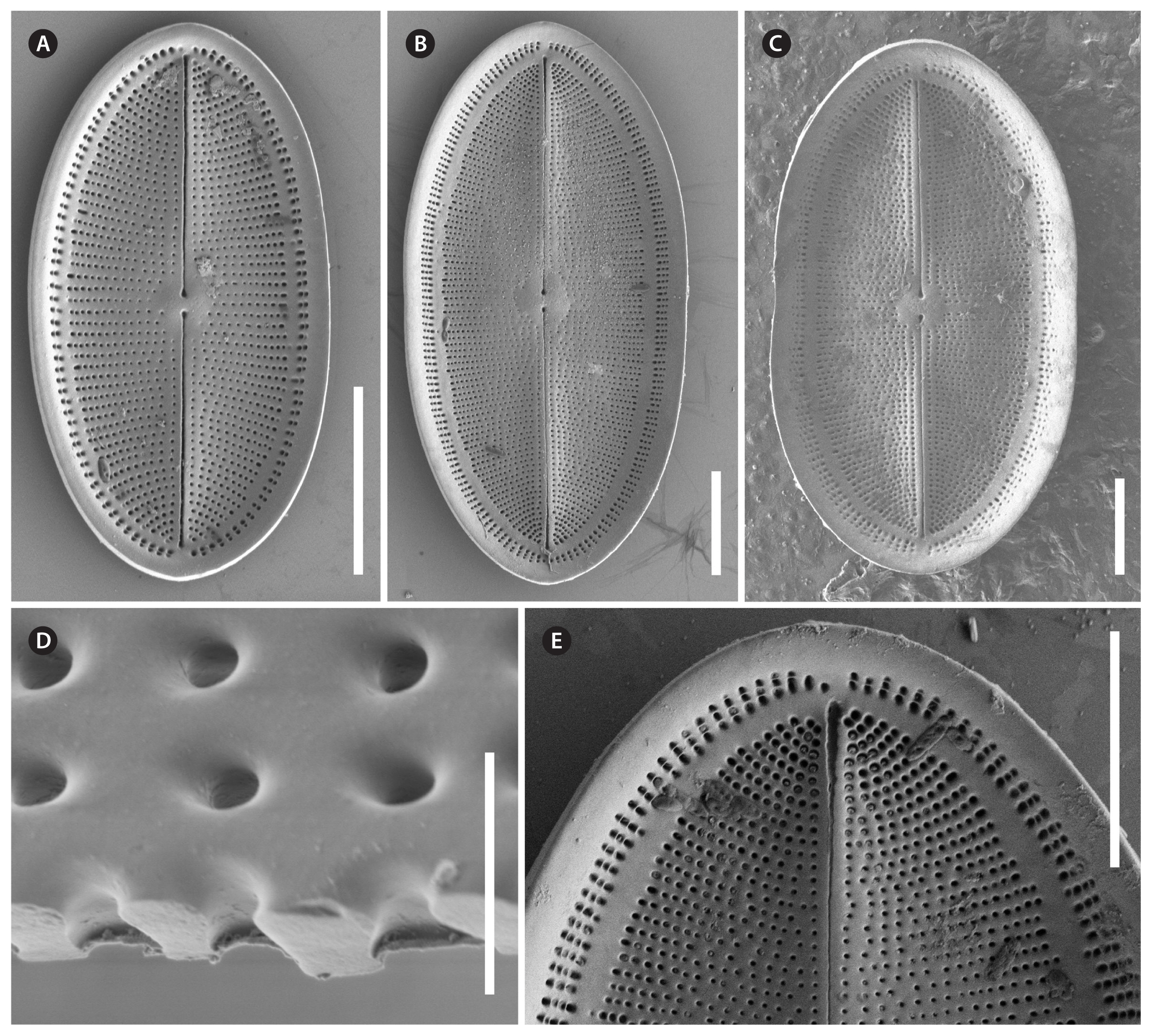
Fig. 8
Cocconeis coreana, strain D119_029, scanning electron microscope. Raphe valve (RV) internal view. (A) RV with open valvocopula and irregularly spaced clamp-like fimbriae seemingly attached to the hyaline rim, and the proximal raphe ends bent to opposite directions; underneath this valve lies a sternum valve (SV) internal view. (B & D) Details of round areolae with a diameter of about 0.25 μm and round lattice-structured hymenes. (C) Large RV without valvocopula, note the heavily silicified hyaline rim; underneath this valve lies a small SV external view. Scale bars represent: A & C, 10 μm; B, 1 μm; D, 0.5 μm. 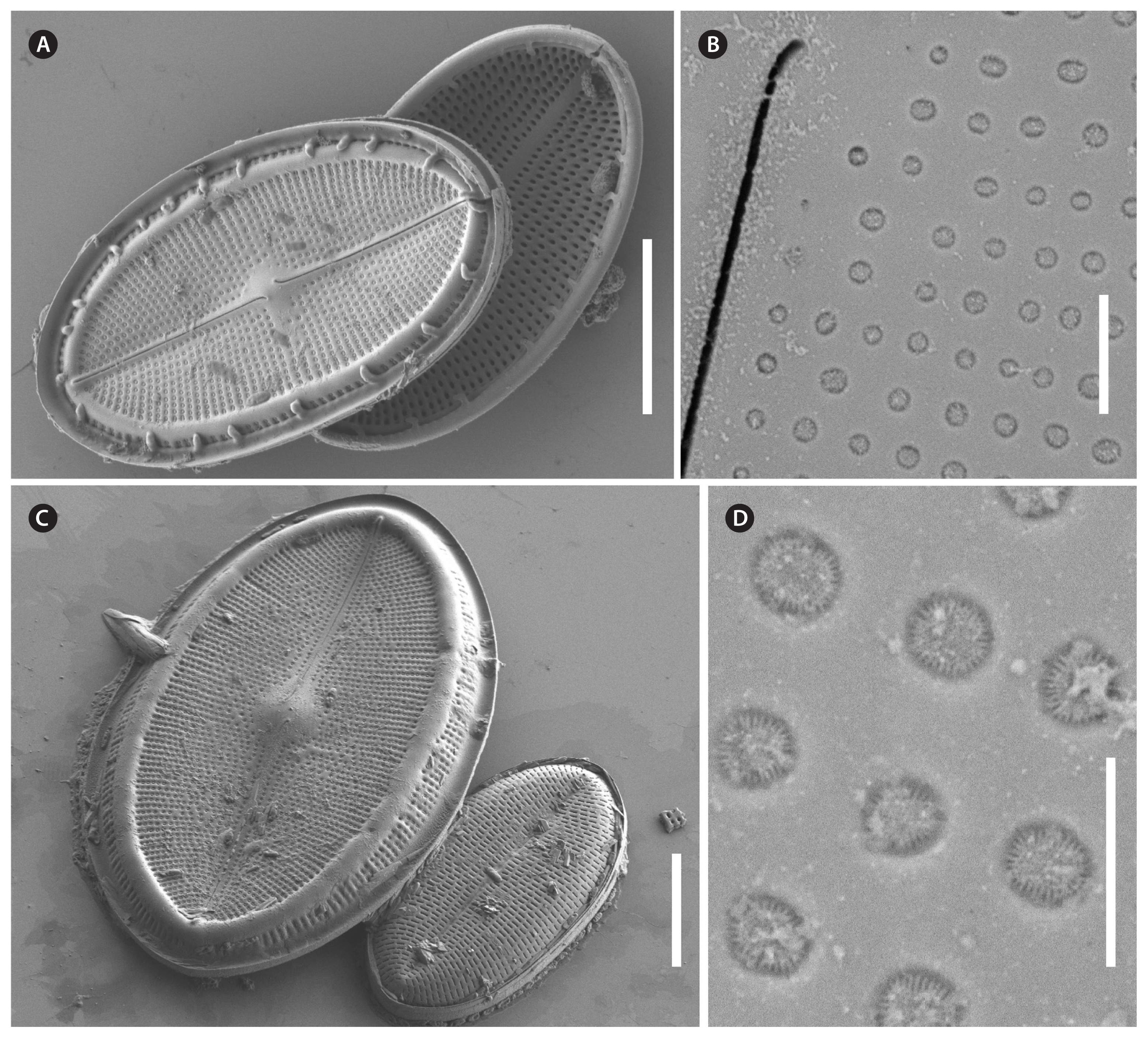
Fig. 9
Cocconeis coreana, scanning electron microscope. Sternum valve (SV) internal views. (A) With no valvocopula. (B) With attached smooth open sternum valvocopula. (C) With attached open fimbriated valvocopula of the raphe valve. (D–G) Details of areolae, open internally and covered by a dash-like hymen externally. (D) Overview. (E) Broken SV with view unto the elongated lattice-structured hymen. (F) Look into the open areolae to the hymen close to the exterior of the valve. (G) Broken areolae with remnants of hymenes. (A), strain K13rj7_9; (B–G), strain D119_029. Scale bars represent: A–C, 10 μm; D–G, 1 μm. 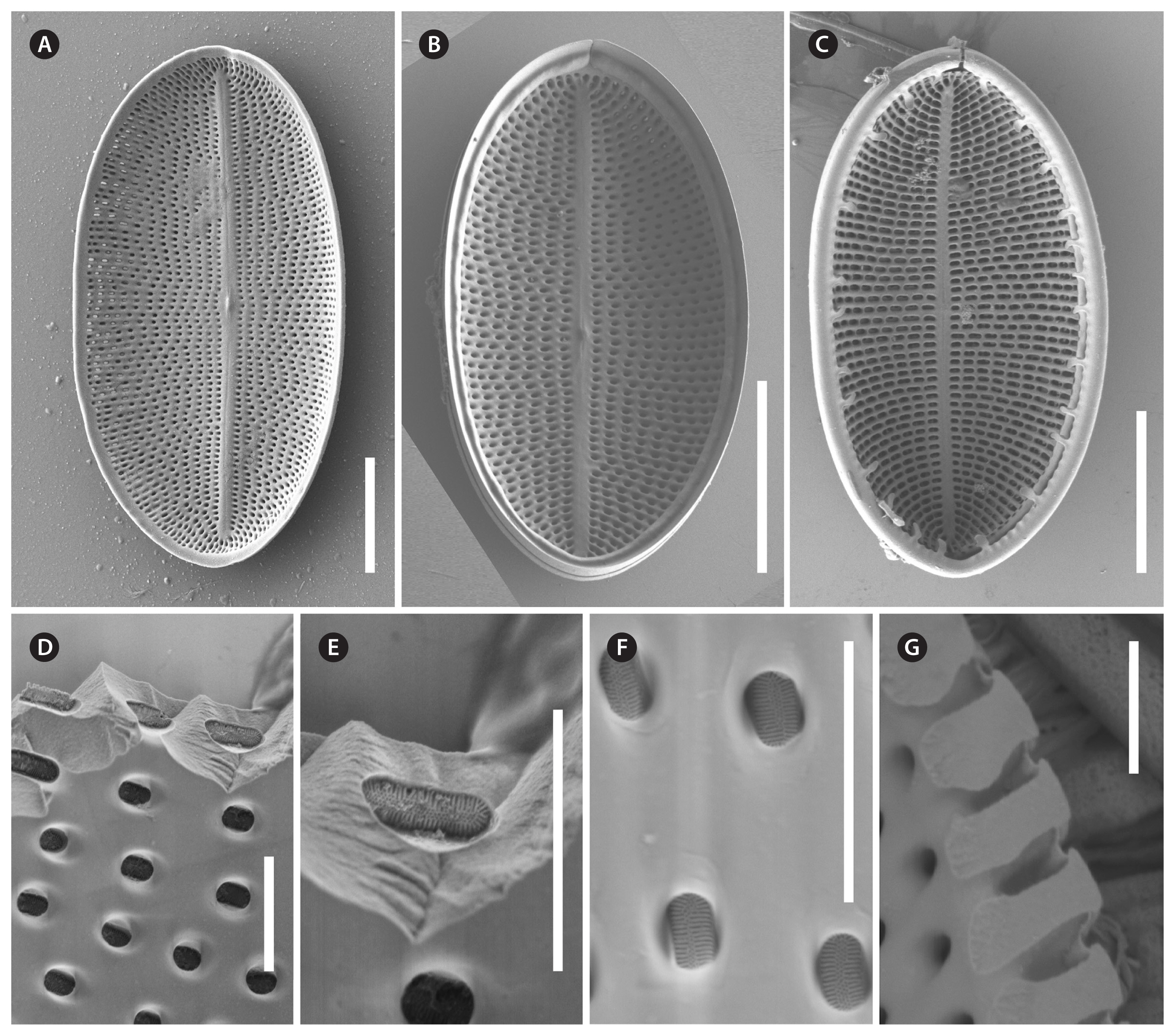
Fig. 10
Cocconeis coreana, scanning electron microscope. (A, B, D & E) Sternum valve (SV) external view with dash-like areolae. Note the different slit sizes in (A) and (D). (A) Note the raphe like imprints on the sternum. (B) Areolae with elongated lattice-structured hymenes (0.7 μm length by 0.13 μm width) close to the exterior of the valve. (C) Smooth open (top) valvocopulae of SV. (F) Fimbriated open (top) valvocopula of raphe valve. (A & B), strain D119_029; (C & F), strain D26_016; (D & E), strain K13rj7_9. Scale bars represent: A, C, D & F, 10 μm; B, 0.5 μm; E, 1 μm. 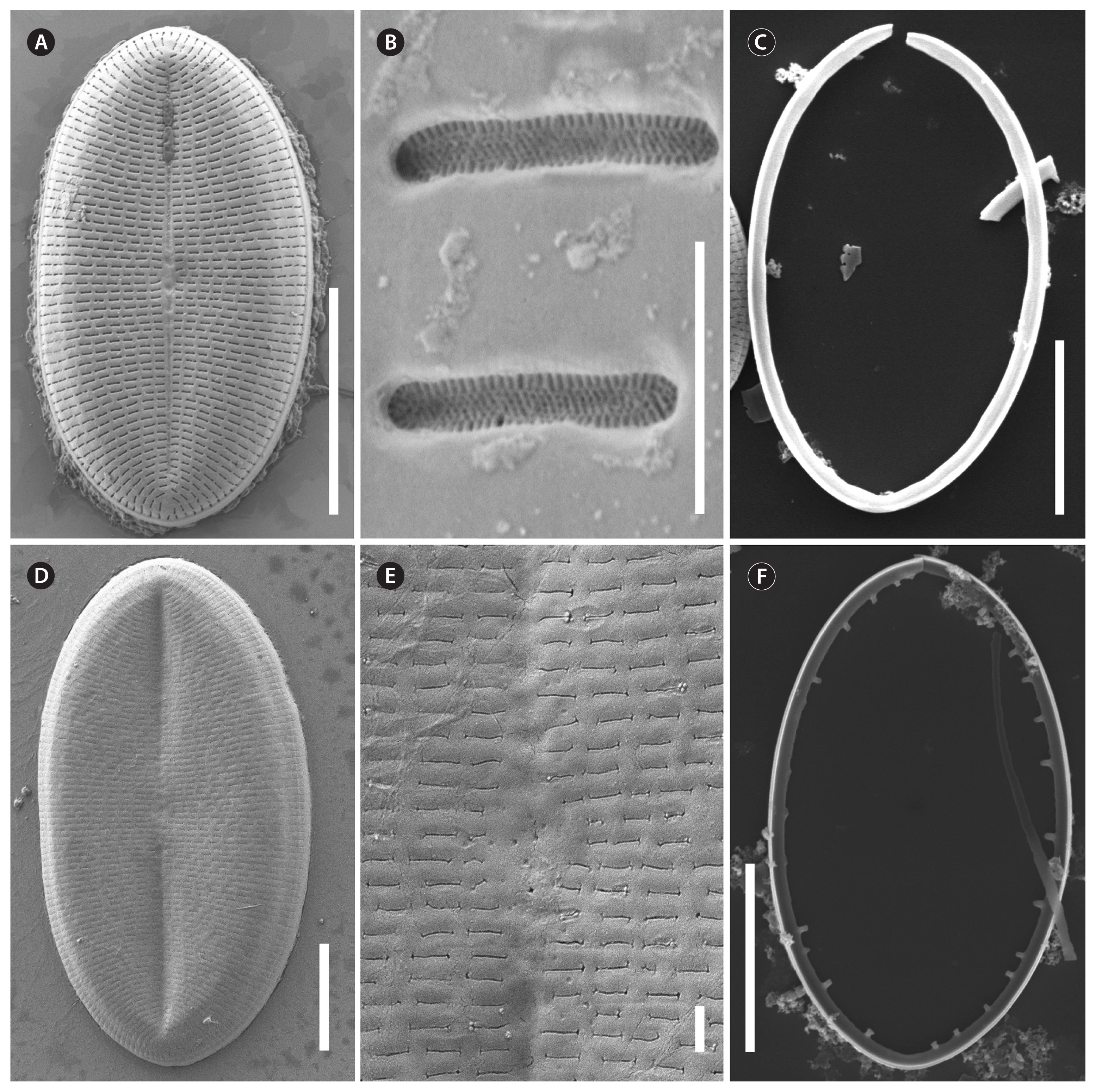
Fig. 11
Cocconeis coreana, strain D119_029, scanning electron microscope. (A–D) Different stages in auxospore formation. (A) Remnants of early auxospore surrounded by incunabula. (B) Domed initial sternum valve, external view with remnants of incunabula still attached. (C) Precursor of initial valve external view. (D) Precursor of initial valve internal view. Scale bars represent: A–D, 10 μm. 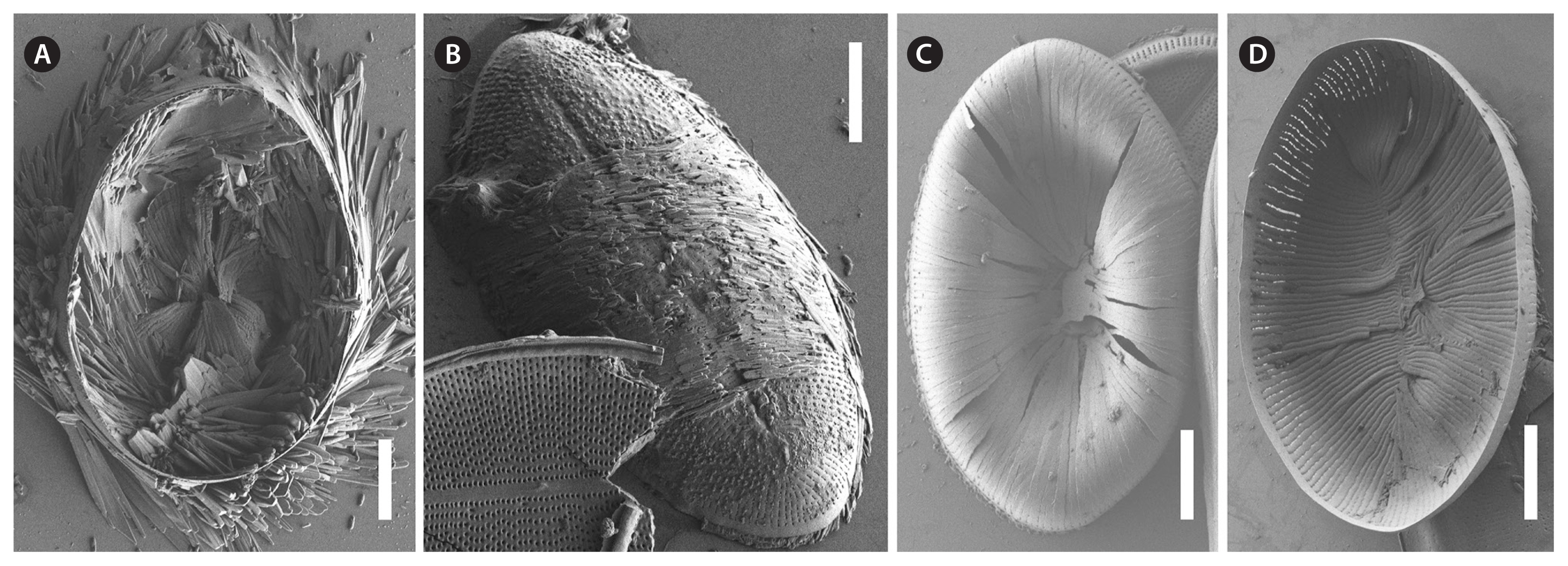
Fig. 12
Cocconeis sijunghoensis, strain NK8_32b (type strain), light microscopy. (A–E) Raphe valve (RV). (F–J) Sternum valve (SV). (F) (SV; Fig. A RV) represents the holotype. Scale bar represents: A–J, 10 μm. 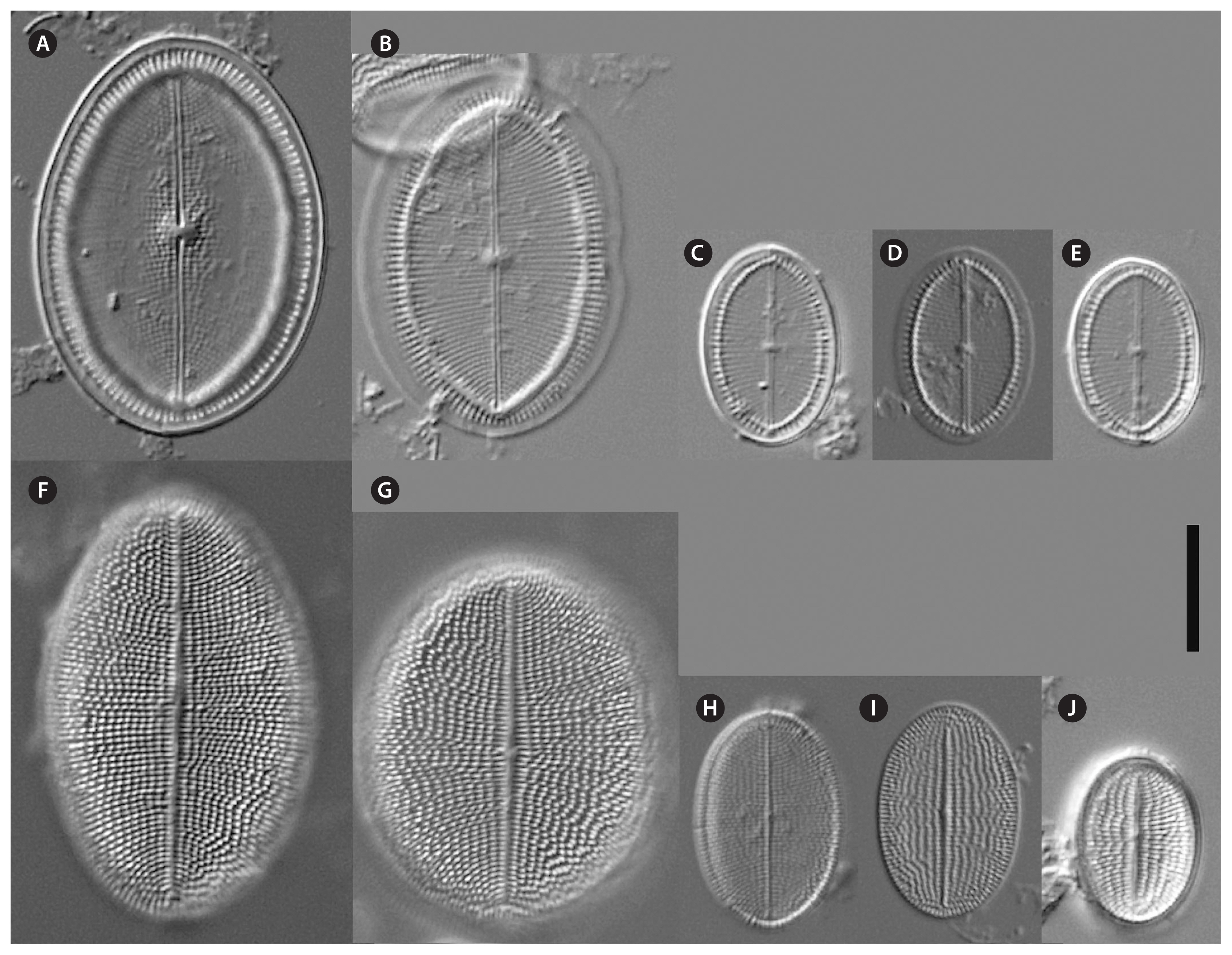
Fig. 13
Cocconeis sijunghoensis, strain NK8_32b, scanning electron microscope. (A–F) Raphe valve (RV). (A & B) External view, note the hyaline rim with only 1–3 areolae between rim and edge. (B) Partly girdle view of complete valve with main view unto the RV. (C) Fimbriated raphe valvocopula. (D–F) RV internal view, note the hyaline rim. (F) Hymenes in the areolae. Scale bars represent: A–E, 10 μm; F, 1 μm. 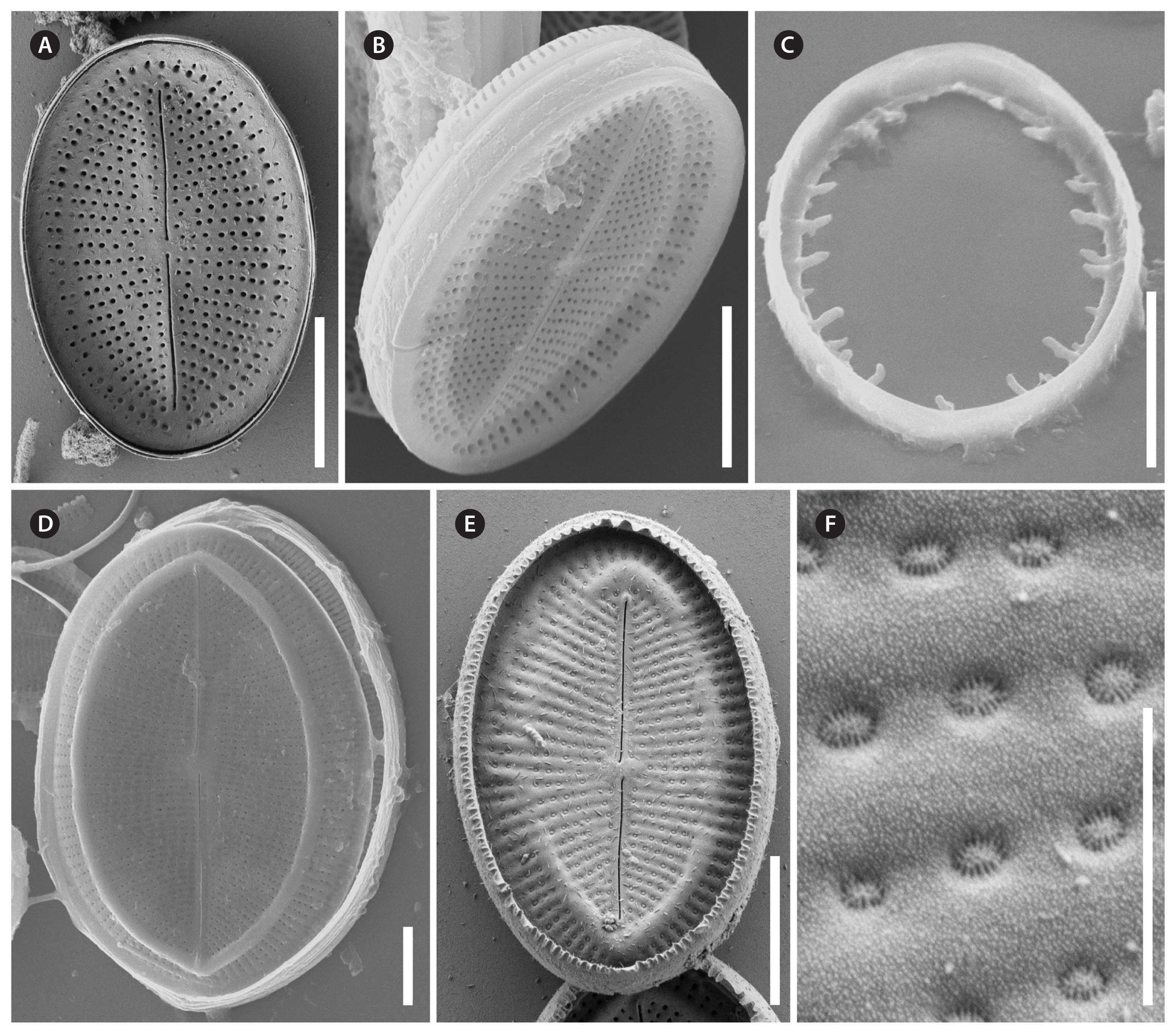
Fig. 14
Cocconeis sijunghoensis, strain NK8_32b, scanning electron microscope. (A–C) Sternum valve (SV). (A & B) External view, note the dash-like areolae covered by elongated hymenes. (C) Internal view, note the lopsidedness of the sternum, and the smooth valvocopula of the SV, on the left side covered by a broken and fimbriated valvocopula of the raphe valve. Scale bars represent: A & C, 10 μm; B, 1 μm. 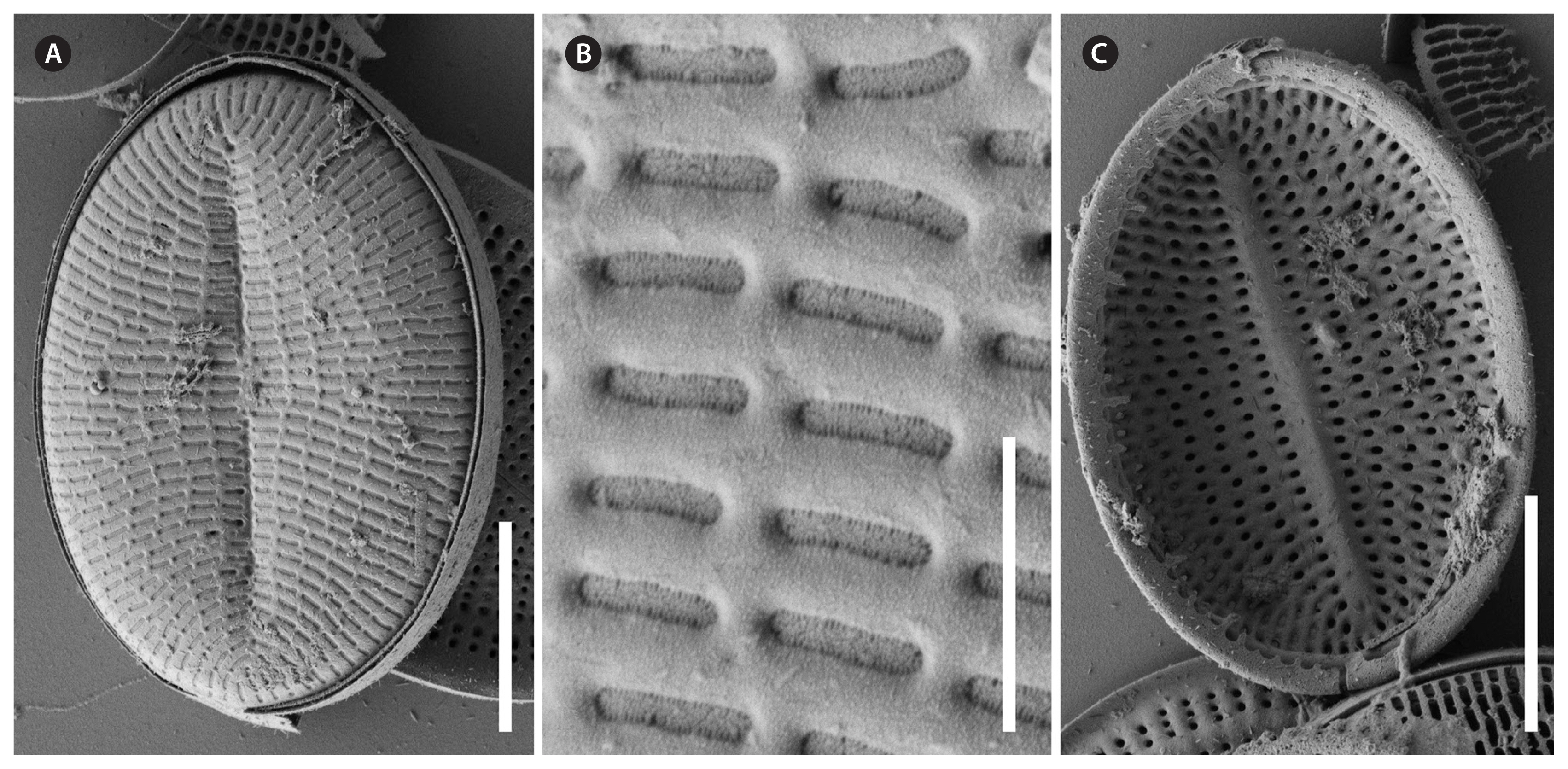
Table 1Taxon name, strain ID, and morphometric data of studied Cocconeis strains
a Corrected data of Romero and Jahn (2013). Table 2Genetic distances (uncorrected p-distances) between rbcL sequences of Cocconeis taxa Table 3Genetic distances (uncorrected p-distances) between 18SV4 sequences of Cocconeis taxa REFERENCESAbarca, N., Jahn, R., Zimmermann, J. & Enke, N. 2014. Does the cosmopolitan diatom Gomphonema parvulum (Kützing) Kützing have a biogeography? PLoS ONE. 9:e86885 pp.
Abarca, N., Zimmermann, J., Kusber, W., Mora, D., Van, AT., Skibbe, O. & Jahn, R. 2020. Defining the core group of the genus Gomphonema Ehrenberg with molecular and morphological methods. Bot Lett. 167:114–159.
Abdel-Rahman, ME. & Sinada, FA. 2016. An updated checklist of the phytoplankton of the White Nile at Khartoum, Sudan. Eur Acad Res. 4:5837–5845.
Al-Handal, AY. & Al-Shaheen, MA. 2019. Diatoms in the wetlands of Southern Iraq. Bibl Diatomol. 67:1–252.
Amutha, M. & Muralidharan, M. 2017. Diatom community structure along physicochemical gradients in upland river segments of Tamiraparani river system, South India. Int J Aquat Biol. 5:12–21.
Chen, S., Zhang, W., Zhang, J., Jeppesen, E., Liu, Z., Kociolek, JP., Xu, X. & Wang, L. 2019. Local habitat heterogeneity determines the differences in benthic diatom metacommunities between different urban river types. Sci Total Environ. 669:711–720.
Costa, LF., Wetzel, CE., Ector, L. & Bicudo, DC. 2020. Freshwater Cocconeis species (Bacillariophyceae) from Southeastern Brazil, and description of C. amerieuglypta sp. nov. Bot Lett. 167:15–31.
Droege, G., Barker, K., Astrin, JJ., Bartels, P., Butler, C., Cantrill, D., Coddington, J., Forest, F., Gemeinholzer, B., Hobern, D., Mackenzie-Dodds, J., Tuama, ÉÓ., Petersen, G., Sanjur, O., Schindel, D. & Seberg, O. 2014. The global genome biodiversity network (GGBN) data portal. Nucleic Acids Res. 42:D607–D612.
Edgar, RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 26:2460–2461.
Gari, EN. & Corigliano, MC. 2007. Spatial and temporal variations of Cocconeis placentula var. euglypta (Ehrenberg.) 1854 Grunow, 1884 in drift and periphyton. Braz J Biol. 67:587–595.
Geitler, L. 1973. Auxospore formation and systematics in pennate diatoms and the cytology of Cocconeis races. Österr. Bot Z. 122:299–321.
Gruenstaeudl, M. 2020. Annonex2embl: automatic preparation of annotated DNA sequences for bulk submissions to ENA. Bioinformatics. 36:3841–3848.
Heinrich, CG., Palacios-Peñaranda, ML., Peña-Salamanca, E., Schuch, M. & Lobo, EA. 2019. Epilithic diatom flora in Cali River hydrographical basin, Colombia. Rodriguésia. 70:e02062017 pp.
Hofmann, G., Werum, M. & Lange-Bertalot, H. 2013. Aufwuchs-Diatomeen im Süβwasser-Benthos von Mitteleuropa. Bestimmungsflora Kieselalgen für die ökologische Praxis. Über 700 der häufigsten Arten und ihre Ökologie. 2. Korrigierte Auflage. A.R.G. Gantner, Ruggell, 908 pp.
Holmes, RW., Crawford, RM. & Round, FE. 1982. Variability in the structure of the genus Cocconeis Ehr. (Bacillariophyta) with special reference to the cingulum. Phycologia. 21:370–381.
Jahn, R., Abarca, N., Gemeinholzer, B., Mora, D., Skibbe, O., Kulikovskiy, M., Gusev, E., Kusber, W-H. & Zimmermann, J. 2017.
Planothidium lanceolatum and Planothidium frequentissimum reinvestigated with molecular methods and morphology: four new species and the taxonomic importance of the sinus and cavum. Diatom Res. 32:75–107.
Jahn, R. & Kusber, W-H. 2005. (continuously updated). AlgaTerra information system, Botanic Garden and Botanical Museum Berlin, Freie Universität, Berlin, Available from: http://www.algaterra.org
. Accessed Mar 27, 2020
Jahn, R., Kusber, W-H. & Romero, OE. 2009.
Cocconeis pediculus Ehrenberg and C. placentula Ehrenberg var. placentula (Bacillariophyta): typification and taxonomy. Fottea. 9:275–288.
Jahn, R., Kusber, W-H., Skibbe, O., Zimmermann, J., Van, AT., Buczkó, K. & Abarca, N. 2019.
Gomphonella olivacea (Bacillariophyceae): a new phylogenetic position for a well-known taxon, its typification, new species and combinations. Plant Ecol Evol. 152:219–247.
Joh, G. 2012. Algal flora of Korea, Vol. 3. No. 7. Freshwater diatoms V, Available from: http://webbook.me.go.kr/DLi-File/099/008/5527836.pdf
. Accessed Mar 27, 2020
Jüttner, I., Bennion, H., Carter, C., Cox, EJ., Ector, L., Flower, R., Jones, V., Kelly, MG., Mann, DG., Sayer, C., Turner, JA. & Williams, DM. 2020. Freshwater diatom flora of Britain and Ireland, Amgueddfa Cymru, National Museum Wales, Available from: https://naturalhistory.museumwales.ac.uk/diatoms
. Accessed Apr 19, 2020
Kaczmarska, I., Lovejoy, C., Potvin, M. & MacGillivary, M. 2009. Morphological and molecular characteristics of selected species of Minidiscus (Bacillariophyta, Thalassiosiraceae). Eur J Phycol. 44:461–475.
Kaczmarska, I., Poulíčková, A., Sato, S., Edlund, MB., Idei, M., Watanabe, T. & Mann, DG. 2013. Proposals for a terminology for diatom sexual reproduction, auxospores and resting stages. Diatom Res. 28:263–294.
Keck, F., Rimet, F., Franc, A. & Bouchez, A. 2016. Phylogenetic signal in diatom ecology: perspectives for aquatic ecosystems biomonitoring. Ecol Appl. 26:861–872.
Kermarrec, L., Franc, A., Rimet, F., Chaumeil, P., Humbert, JF. & Bouchez, A. 2013. Next-generation sequencing to inventory taxonomic diversity in eukaryotic communities: a test for freshwater diatoms. Mol Ecol Resour. 13:607–619.
Kobayasi, H., Idei, M., Mayama, S., Nagumo, T. & Osada, K. 2006. H. Kobayasi’s Atlas of Japanese diatoms based on electron microscopy. Uchida Rakakuho Publishing, Tokyo, 531 pp.
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
Lange-Bertalot, H., Hofmann, G., Werum, M. & Cantonati, M. 2017. Freshwater benthic diatoms of Central Europe: over 800 common species used in ecological assessment. Koeltz Botanical Books, Schmitten-Oberreifenberg, 942 pp.
Leterme, SC., Ellis, AV., Mitchell, JG., Buscot, M., Pollet, T., Schapira, M. & Seuront, L. 2010. Morphological flexibility of Cocconeis placentula (Bacillariophyceae) nanostructure to changing salinity levels. J Phycol. 46:715–719.
Mann, DG., McDonald, SM., Bayer, MM., Droop, SJM., Chepurnov, VA., Loke, RE., Ciobanu, A. & Hans du Buf, JM. 2004. The Sellaphora pupula species complex (Bacillariophyceae): morphometric analysis, ultrastructure and mating data provide evidence for five new species. Phycologia. 43:459–482.
Metzeltin, D., Lange-Bertalot, H. & Nergui, S. 2009. Diatoms in Mongolia. Iconographia Diatomologica. 20:Gantner Verlag, Ruggel, 691 pp.
Mizuno, M. 1998. Sexual reproduction and auxospore formation of the marine monoraphid diatom Cocconeis pellucida
. Diatom Res. 13:103–112.
Mora, D., Carmona, J., Jahn, R., Zimmermann, J. & Abarca, N. 2017. Epilithic diatom communities of selected streams from the Lerma-Chapala Basin, Central Mexico, with the description of two new species. PhytoKeys. 88:39–69.
Müller, J., Müller, K., Neinhuis, C. & Quandt, D. 2010. PhyDE: phylogenetic data editor. Computer program. Version 0.9971, Available from: http://www.phyde.de
. Accessed Oct 26, 2016
Ouyang, L., Pan, Y., Huang, C., Tang, Y., Du, J. & Xiao, W-Y. 2016. Water quality assessment of benthic diatom communities for water quality in the subalpine karstic lakes of Jiuzhaigou, a world heritage site in China. J Mt Sci. 13:1632–1644.
PhycoBank. 2017. (continuously updated). Registration of nomenclatural acts of algae, Botanic Garden and Botanical Museum Berlin, Available from: https://www.phycobank.org/
. Accessed Mar 29, 2020
Plenković-Moraj, A., Kralj, L. & Gligora, M. 2008. Effect of current velocity on diatom on colonization glass slides in unpolluted headwater creek. Period Biol. 110:291–295.
Potapova, M. & Spaulding, S. 2013. Diatoms of North America.
Cocconeis placentula sensu lato, Available from: https://diatoms.org/species/cocconeis_placentula
. Accessed Apr 19, 2020
Poulićkova, A., Veselá, J., Neustupa, J. & Skaloud, P. 2010. Pseudocryptic diversity versus cosmopolitanism in diatoms: a case study on Navicula cryptocephala Kütz. (Bacillariophyceae) and morphological similar taxa. Protist. 161:353–369.
Pumas, C., Pruetiworanan, S. & Peerapornpisal, Y. 2018. Diatom diversity in some hot springs of northern Thailand. Botanica. 24:69–86.
Rasband, WS. 1997–2016. ImageJ. Image processing and analysis in Java, Available from: https://imagej.nih.gov/ij/
. Accessed Mar 29, 2020
Romero, OE. & Jahn, R. 2013. Typification of Cocconeis lineata and Cocconeis euglypta (Bacillariophyta). Diatom Res. 28:175–184.
Ruck, EC. & Theriot, EC. 2011. Origin and evolution of the canal raphe system in diatoms. Protist. 162:723–737.
Skibbe, O., Zimmermann, J., Kusber, W-H., Abarca, N., Buczkó, N. & Jahn, R. 2018.
Gomphoneis tegelensis sp. nov. (Bacillariophyceae): a morphological and molecular investigation based on selected single cells. Diatom Res. 33:251–262.
Stancheva, R. 2019.
Cocconeis cascadensis, a new monoraphid diatom from mountain streams in Northern California, USA. Diatom Res. 33:471–483.
Taylor, JC. & Cocquyt, C. 2016. Diatoms from the Congo and Zambezi basins: methodologies and identification of the genera. ABC Taxa and the Belgian Development Cooperation, Brussels, 364 pp.
Toudjani, AA., Çelekli, A., Gümüs, EY., Kayhan, S., Lekesiz, HÖ. & Çetin, T. 2017. A new diatom index to assess ecological quality of running waters: a case study of water bodies in western Anatolia Turkey. Ann Limnol Int J Limnol. 53:333–343.
Turland, NJ., Wiersema, JH., Barrie, FR., Greuter, W., Hawksworth, DL., Herendeen, PS., Knapp, S., Kusber, W-H., Li, D-Z., Marhold, K., May, TW., McNeill, J., Monro, AM., Prado, J., Price, MJ. & Smith, GF. 2018. In: International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress; Shenzhen, China. July 2017; Koeltz Botanical Books, Glashütten, 254 pp.
Wetzel, CE., Beauger, A. & Ector, L. 2019.
Cocconeis rouxii Héribaud & Brun a forgotten, but common benthic diatom species from the Massif Central, France. Bot Lett. 166:221–233.
Wortley, AH. & Scotland, RW. 2006. The effect of combining molecular and morphological data in published phylogenetic analyses. Syst Biol. 55:677–685.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||