ABSTRACTOcean warming can have significant negative impacts on population genetic diversity, local endemism and geographical distribution of a wide range of marine organisms. Thus, the identification of conservation units with high risk of extinction becomes an imperative task to assess, monitor, and manage marine biodiversity for policy-makers. Here, we surveyed population structure and genetic variation of the red seaweed Gracilaria vermiculophylla along the coast of China using genome-based amplified fragment length polymorphism (AFLP) scanning. Regardless of analysis methods used, AFLP consistently revealed a south to north genetic isolation. Populations at the southern coast of China showed unique genetic variation and much greater allelic richness, heterozygosity, and average genetic diversity than the northern. In particular, we identified a geographical barrier that may hinder genetic exchange between the two lineages. Consequently, the characterized genetic lineage at the southern coast of China likely resulted from the interplay of post-glacial persistence of ancestral diversity, geographical isolation and local adaptation. In particular, the southern populations are indispensable components to explore evolutionary genetics and historical biogeography of G. vermiculophylla in the northwestern Pacific, and the unique diversity also has important conservation value in terms of projected climate warming.
INTRODUCTIONGlobal climate warming has become a serious threat to the entire marine ecosystems on the earth. It was estimated that the global mean sea surface temperature (SST) increased by approximately 0.4°C since the 1950s due to most of the global heat being absorbed by the upper layers of the oceans (Doney et al. 2012). Consequently, the warming SST altered stratification of water column, oxygen content, nutrient availability and primary production, and ultimately changed diversity gradients of a wide variety of marine taxa (Ramírez et al. 2017). Recent phylogeographical studies illustrated the contraction of geographical range and biomass loss of seaweed species in the East Atlantic driven by ocean warming, including some genetic lineages with extinction risk at the rear-edge of the range (Provan and Maggs 2012, Neiva et al. 2015). At the same time, extreme climate events (e.g., heat waves) were reported to dislodge or eradicate foundation species in marine ecosystems, subsequently a striking shift in community structure towards a depauperate state (Wernberg et al. 2013). It is thus an imperative task to identify genetic pools and / or geographical regions with high conservation values to mitigate biodiversity loss under projected ocean warming (Pauls et al. 2013).
The red seaweed Gracilaria vermiculophylla (Ohmi) Papenfuss was initially described from Hokkaido, Japan (as Gracilariopsis vermiculophylla), and it had subsequently been documented throughout the Northwestern Pacific (Tseng and Xia 1999, Terada and Yamamoto 2002, Kim et al. 2010). This agar-producing red species usually occurs in the intertidal zone and sometimes luxuriantly in shallow estuaries (Terada and Yamamoto 2002). Ecologically, G. vermiculophylla plays an essential role in structuring and maintaining costal marine communities (Ramus et al. 2017). For instance, recent experimental manipulations and ecological interactions showed that G. vermiculophylla can provide biomass-dependent multiple ecological functionalities in localized coastal marine ecosystems, including abundance, richness, diversity, and density of nursery taxa (Ramus et al. 2017). In addition, G. vermiculophylla has also been used as an efficient component of land-based Integrated Multi-Trophic Aquaculture (IMTA) systems to mitigate environmental problems caused by fed aquaculture (Abreu et al. 2011).
The past decades have witnessed G. vermiculophylla to be a notorious marine invader to colonize the east Pacific, the Atlantic and the Baltic Sea, the Mediterranean Sea and Morocco (Kim et al. 2010, Gorman et al. 2017, Krueger-Hadfield et al. 2017). Multiple lines of evidence, including recent genetic phenotyping, indicate that some unique ecological traits (e.g., tolerance to grazing, extreme salinities and temperature, reviewed in Hu and Juan 2014) and microevolution to climatic niche shift in the introduced habitats account for the rapid global invasion of G. vermiculophylla (Sotka et al. 2018). Meanwhile, a few studies tried to explore the donor region of the invaded populations. For example, Kim et al. (2010) used mitochondrial cox1 sequencing and first reported the East Sea / Sea of Japan as a potential source of invaded G. vermiculophylla in the eastern Pacific and the Atlantic. However, recent genetic analysis indicated that northeastern Japan more likely served as the donor region than other native areas (Krueger-Hadfield et al. 2017).
G. vermiculophylla is widely distributed in China with range from the northern Dalian, Liaoning to the southern BeiHai, Guangxi (Tseng and Xia 1999) (Fig. 1). No historical evidence indicate G. vermiculophylla occurring in Hainan Island (Fig. 1), and our recent year-rounds of surveys support the absence. Previous studies of G. vermiculophylla in China mainly focused on temperature-induced growth, development and propagation (Tseng and Chen 1959), and aquaculture and mitigating effects on eutrophication (Xu et al. 2008). Recently, Liu et al. (2016) studied population genetic diversity of G. vermiculophylla along the coast of China using mitochondrial cox1. However, the reasons contributing to geographical isolation and fine-scale population genetic characteristics have not been addressed, which limits our understanding of how G. vermiculophylla established the present-day biogeographical patterns throughout the native range.
In this study, we used amplified fragment length polymorphism (AFLP)-based genome scanning to survey 18 G. vermiculophylla populations along the coast of China. Our goal was to characterize fine-scale population genetic variation and examine if there is a geographical barrier contributing to lineage isolation. These knowledge should help us guide the collection and ex situ conservation of unique diversity of G. vermiculophylla under increasing climate warming and human impacts.
MATERIALS AND METHODSSpecimens sampling and DNA extraction
G. vermiculophylla specimens were collected from 18 sites ranging from Dalian, Liaoning (122.74° N, 39.04° E) to Beihai, Guangxi (109.34° N, 21.45° E) between 2013 and 2015 (Table 1, Fig. 1). We treated the five sites (YT, JH, TC, NA, and ZP) close to Guangxi, China as the southern coast and the other thirteen sites as the northern coast (Fig. 1). At each site, specimens were collected with an interval transect ≥ 10 m to avoid sampling related plants. In total, we obtained 456 individuals along the coast of China. All specimens were preserved with silica gel. Genomic DNA was extracted following Hu et al. (2015). The purity and quality of genomic DNA were checked by the ratios of A260/A280 and visualized by gel electrophoresis.
AFLP genome scanningAFLP analysis followed Vos et al. (1995). Digest reactions were conducted in a 25 μL of mixture containing 200 ng of DNA, 1× NEBuffer, 8 U EcoRI (New England BioLabs, Ipswich, MA, USA), and 8 U MseI (New England BioLabs). Reactions were incubated for 6 h at 37°C, followed by 20 min at 65°C to denature the enzymes. After incubation, 10 μL of the reaction was run on 1% agarose gels to check the size of digested DNA fragments (250–2,000 bp). Adaptors were ligated to the digested fragments in reactions containing 20 μL of digested DNA, 1× T4 ligation buffer (with ATP), 0.125 μM EcoRI adaptor, 1.25 μM MseI adaptor, and 175 U of T4 DNA ligase (New England BioLabs). Reactions were incubated for 8 h at 16°C.
Pre-amplifications were performed in a 20 μL of mixture containing 0.5 μL of digested and ligated DNA, 4 μM EcoRI + A primer, 4 μM MseI + C primer, 1× EasyTaq Buffer, 3.75 mM of dNTPs, and 2 U EasyTaq DNA polymerase. Polymerase chain reaction (PCR) reactions consisted of an initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 1 min, and a final extension at 72°C for 5 min. Ten μL of pre-amplification product was visualized on 1% agarose gels to check the size of visible smears (250–750 bp). The remaining 10 μL was diluted 1 : 20 by adding TE buffer.
Selective amplifications were performed with EcoRI and MseI primers containing three selective nucleotides (Supplementary Table S1). The PCR was conducted in a 20 μL of mixture containing 0.5 μL of diluted pre-amplification product, 4 μM EcoRI primer, 4 μM MseI primer, 1× EasyTaq Buffer, 3.75 mM of dNTPs, and 2 U EasyTaq DNA polymerase. Reactions were carried out in an Eppendorf Mastercycler Nexus Gradient (Eppendorf, Hamberg, Germany) using the following profiles: 5 min at 94°C, 12 cycles of 30 s at 94°C, 30 s at 65–0.7°C/cycle after the first cycle and 60 s at 72°C, followed by 25 cycles of 30 s at 94°C, 60 s at 56°C and 60 s at 72°C, with a final extension of 5 min at 72°C. In total, five selective amplifications were conducted to screen all 456 individuals of G. vermiculophylla. Products were run on 5% acrylamide gels and finally five primer combinations (Supplementary Table S1) were determined based on the number of bands, polymorphisms and visible sizes (150–500 bp). Products were then loaded onto an ABI 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) for capillary electrophoresis with the GeneScan―500 LIZ (Applied Biosystems) size standard. To test the reproducibility of AFLP analyses in G. vermiculophylla, we randomly chose two specimens from each population and examined three replicates per individual. The differences between replicates for each individual assayed by the five primer combinations were counted and divided by the total number of phenotypic comparisons to obtain the error rate (Bonin et al. 2004).
Genetic diversity and population structureGeneMarker v2.2.0 (SoftGenetics, State College, PA, USA) was used to allocate the binary data to appropriate loci. After pre-analysis using default settings, size profiles of all samples were checked and corrected manually. AFLPScore v1.4 (Whitlock et al. 2008) was used to call peak height data and minimize error whilst maximizing the number of retained loci by setting a locus selection threshold of 5% (the average intensity of bands at a specific locus, above which a locus is retained in the dataset). The exported phenotype matrix was analyzed using GenAlEx 6.501 (Peakall and Smouse 2012) to obtain basic diversity indices, including proportion of polymorphic loci (PPL), observed number of alleles (Na), effective number of allele (Ne), Shannon’s information index (I), unbiased expected heterozygosity (uHe), principal component analysis (PCA), and analysis of molecular variance (AMOVA). Two other indices, average gene diversity over loci (Hd) and number of allele (Na), and population-pairwise FST values were obtained using Arlequin 3.5.1.2 (Excoffier and Lischer 2010) with 1,000 iterations. Arlequin was used to perform partition of genetic variation in G. vermiculophylla and compare the consistency relative to the program GenAlEx. Genetic differentiation was also measured with Jost’s D distance between each site using Spade (Chao and Shen 2003). Unweighed Pair Group Method with Arithmetic mean (UPGMA) dendrogram was also constructed to measure genetic relationships of all 456 individuals of G. vermiculophylla.
To visualize the pattern of genetic clustering of populations, we used STRUCTURE 2.3.4 (Pritchard et al. 2000) to assess genetic relatedness of 18 populations. We ran STRUCTURE by setting the number of predefined groups (K = 1–5) and a model based on admixture and independent allelic frequencies. Each run consisted of 106 Markov Chain Monte Carlo (MCMC) repetitions with a burn-in period of 105 MCMC repetitions. The analysis was run for 10 replicates to allow averaging of results in CLUMPP 1.1.2 (Jakobsson and Rosenberg 2007). The graphical representation of STRUCTURE results was finally plotted in DISTRUCT 1.1 (Rosenberg 2004).
Biogeographical barriersTo detect potential biogeographical barriers contributing to population genetic isolation in G. vermiculophylla along the coast of China, we conducted a post hoc spatial analysis using Barrier 2.2 (Manni et al. 2004). Barrier characterizes a computational geometry using Voronoi tessellation and Delaunay triangulation to visualize patterns of genetic variation. Genetic distances were mapped onto a matrix of geographical coordinates of sampling sites, and their spatial relationships were modeled to determine borders between neighboring sites that had the highest levels of genetic differentiation and ranked them accordingly (Manni et al. 2004). We tested the robustness of the computed barriers using re-sampled bootstrapped pairwise FST distances, performing 10 replicate analyses.
RESULTSAFLP profiles showed good reproducibility in G. vermiculophylla with a mean error rate of 2.9% across all replicated samples. Five primer combinations produced a total of 987 loci in the range of 60–498 bp for 18 populations, of which 434 were polymorphic (43.97%) (Supplementary Table S1). AFLP results also showed a high proportion of polymorphic loci (mean = 63.41%) and rich number of private AFLP bands (mean = 8.4) in the southern YT, JH, TC, NA, and ZP populations, while the corresponding values for the northern thirteen populations were 37.35% and 1.69, respectively (Supplementary Table S2).
The measurements of PPL, Na, Ne, I, uHe, Hd, and Na (Table 1) showed that the maximum diversity values occurred in the five southern populations (YT, JH, TC, NA, and ZP), whereas the populations from the northern coast had lower genetic values and the minimum estimates were detected in Dalian, Liaoning (Table 1, Fig. 1A). When populations were grouped, the southern coast also showed higher genetic diversity than the northern (Table 1). The spatial analysis of pairwise FST matrix in Barrier identified two physical barriers along the coast of China (Fig. 1B). The two strongly-supported barriers with segment distance of 0.430 and 0.355, respectively, were labeled with Roman numerals in Fig. 1B. For each time of analysis, the barrier between the populations ZP and SS (Fig. 1B) was always the first to be placed regarding of number of designated barriers. The second barrier was found between the populations JM and ZZ across the Bohai Strait (Fig. 1B).
Pairwise FST and Jost’s D distances revealed high genetic differentiation (FST: the mean = 0.401; Jost’s D: the mean = 0.201) between the southern five and the northern 13 populations, and all FST estimates were statistically significant (Table 2). In addition, FST and Jost’s D indicated that the mean genetic differentiation among the southern five populations were 0.067 and 0.050, respectively, while the corresponding estimates among the northern thirteen populations were 0.198 and 0.084, respectively (Table 2). UPGMA dendrogram based on genetic similarity revealed two major clusters in which one cluster (red) comprised five southern populations (YT, IH, TC, NA, and ZP), whereas the northern cluster was further subdivided into two clades (purple and blue) (Fig. 1A). PCA results showed the same population genetic connectivity as UPGMA (Fig. 2A). STRUCTURE analysis gave the highest value of ΔK for K = 2 (Supplementary Fig. S1), it also revealed two distinct genetic groups in G. vermiculophylla (Fig. 2B). Interestingly, a signal of admixture was detected when the K was set to three (Fig. 2B). Locus by locus AMOVA analysis, regardless of using GenALEx or Arlequin, showed that 35% of the variation occurred among regions (south vs. north) and as much as 55% of the variation occurred within populations, only about 10% of the variation was among populations (Supplementary Table S3). In addition, AMOVA results indicated significant genetic structure between the southern and northern coasts (FCT = 0.35, p < 0.0001) (Supplementary Table S3).
DISCUSSIONPopulation genetic differentiation
G. vermiculophylla is a commercially important red seaweed with a long cultivation history in China. Deciphering population genetic structure and diversity patterns can provide important insights for collecting and preserving natural resources and ultimately sustainable selective breeding of the species. In this study, AFLP datasets revealed a south to north genetic break in G. vermiculophylla along the coast of China, a pattern that has also been reported in the brown algae Sargassum hemiphyllum (Cheang et al. 2010), S. horneri (Hu et al. 2011), S. fusiforme (Hu et al. 2017), and S. thunbergii (Li et al. 2017). In the western Pacific ocean, the China Seas consist of three marginal seas (Yellow-Bohai Sea, East China Sea, and South China Sea) (Fig. 1). During the Pleistocene ice periods, the sea-level drops induced by glaciations caused the East China Sea and South China Sea to be enclosed marginal seas, and the Yellow-Bohai Sea was entirely exposed (Wang 1999). As a consequence, we can speculate that the ancestral populations of G. vermiculophylla may have survived in two isolated glacial refugia when sea-levels dropped. One was likely located in the north of South China Sea (e.g., Hainan Island) (Hu et al. 2018) and the surviving ancestral relics subsequently evolved into the present-day southern lineage through post-glacial expansion. The other probably distributed in Okinawa Trough (He et al. 2010, Hu et al. 2011, 2015, 2017) and the ancestral relics re-colonized northward into the Yellow-Bohai Sea when sea-level rose, leading to a significant genetic break along the coast of China. Such a post-glacial population expansion scenario is supported by AFLP-based dendrogram topology (Fig. 1A) (Emerson et al. 2001) and star-like mtDNA cox1 haplotype network reported by Liu et al. (2016).
Interestingly, G. vermiculophylla exhibited a fine-scale population genetic subdivision in Qingdao, Shandong peninsula (Figs 1A & 2). This pattern has never been reported in other seaweeds, including the recently surveyed red alga Chondrus ocellatus (Hu et al. 2015) in which genotypes identified in Qingdao exhibited close relationships to those in the southwest of Japan. Microgeographical environmental variables (e.g., tide height) are likely responsible for such a population subdivision (Krueger-Hadfield et al. 2013). Qingdao, geographically neighboring to the Yellow Sea (Fig. 1), has a temperate monsoon climate that leads to intertidal G. vermiculophylla exposure to semidiurnal tide all over the year. The tidal strength between high- and low-shore stands may exert selection pressure to G. vermiculophylla and hence a microgeographical scale genetic differentiation (Krueger-Hadfield et al. 2013). Meanwhile, G. vermiculophylla in Qingdao occurs in ecologically distinct habitats such as wave-exposed reefs, sheltered rock pools and sandy beaches, which can potentially create genetic variation through selection and adaptation (Zardi et al. 2013). In addition, maritime transportation can also lead to population admixture of G. vermiculophylla between Qingdao and adjoining areas (e.g., southwest Japan) through ballast water or hitching a rid on a ship (Krueger-Hadfield et al. 2013), but this hypothesis needs to be tested by extensive sampling from Korea and Japan (e.g., Kyushu and Shikoku).
Unique genetic diversity at the southern rangePopulation genetic characteristics usually show different patterns across the species’ range. Olsen et al. (2004) reported lower genetic diversity in the edge populations of the eelgrass Zostera marina compared to the central distribution range. However, no geographical variation in genetic diversity was found when the southern edge populations were pooled (Diekmann and Serrão 2012). In this study, the southern G. vermiculophylla, no matter whether they were treated as a whole or not, had much higher intra-populations genetic diversity, private and allelic richness, and much lower inter-population genetic divergence relative to the northern coast of China. These results found in G. vermiculophylla, along with similar diversity patterns reported in the Atlantic seaweeds Chondrus crispus (Provan and Maggs 2012), Fucus ceranoides (Neiva et al. 2012), and F. guiryi (Zardi et al. 2015), contradict the prediction that populations residing at the low-latitude margin are genetically depleted and hence reduced fitness and adaptive capacity (Eckert et al. 2008). The unique diversity pattern of G. vermiculophylla at the southern edge may result from a large subset of ancestral relics survived during the Pleistocene ice ages, because post-glacial recolonization and founder events can influence the distribution of genetic diversity across species’ range to a greater extent than contemporary processes such as habitat fragmentation and contraction (Hampe and Petit 2005). In addition, the Changjiang diluted water (31° N, 121° E) can serve as a physical barrier and profoundly reduce seawater salinities, pH and long-distance dispersal of intertidal marine organisms (Wang et al. 2003). Our biogeographical barrier results (Fig. 1B) indicate that the Changjiang diluted water likely impeded gene flow between the southern and northern G. vermiculophylla populations along the coast of China.
Populations at the range-edge are expected to have small size and restricted to isolated habitats owing to harsher environments, higher isolation and selection (Hampe and Petit 2005, Eckert et al. 2008). Indeed, we found fragmented habitats of G. vermiculophylla at Guangxi Province during field surveys, but the population sizes are not as small as expected. This demographical maintenance may stem from non-hierarchical trade-off between life history strategies and environmental determinants (Nicastro et al. 2010). Populations at the range-edge are also more likely under threat of extinction from climate change and human impacts (Gibson et al. 2009). It is therefore noteworthy to compare eco-physiological and phenological performances between the southern and northern lineages, including reproductive allocation, recruitment rates and acclimation to climate extremes, to identify valuable conservation units with specific tolerance or susceptibility to changing environmental variables (e.g., temperature and desiccation). In addition, selection-mediated adaptation to local environment can result in the development of distinct ecotypes in rear-edge populations (Castric and Bernatchez 2003), similarly to the rich private alleles observed in the southern five populations of G. vermiculophylla (Supplementary Table S2). The unique genetic diversity thus have important conservation implications, particularly considering that these postglacial expanded populations did not recolonize the northern glacial areas in the Northwestern Pacific (Hampe and Petit 2005). Ex situ conservation of germplasm banks of southern G. vermiculophylla populations is feasible in practice and will contribute considerably to ecological restoration of coastal ecosystems and sustainable marine cultivation in China.
SUPPLEMENTARY MATERIAL
Supplementary Table S1. The five primer combinations of AFLP and associated polymorphisms in Gracilaria vermiculophylla along the coast of China (http://www.e-algae.org).
Supplementary Table S2. Analysis of polymorphic loci of Gracilaria vermiculophylla populations along the coast of China (http://www.e-algae.org).
Supplementary Table S3. Locus by locus AMOVA of Gracilaria vermiculophylla populations along the coast of China (http://www.e-algae.org).
Supplementary Fig. S1. Delta K values of the K values tested (left) and mean Ln likelihood of K values tested (right) (http://www.e-algae.org). ACKNOWLEDGEMENTSWe thank Dr. Zhong-Min Sun (IOCAS) and Ms. Xiao-Ling Dai (Guangxi University) for providing seaweed samples or assistance in field collections. Special thank goes to Dr. William Stewart Grant for his critical comments to improve the manuscript. This study was supported by the joint research project between National Natural Science Foundation of China and Thailand Research Fund (41761144057), Sino-German Centre for Research Promotion (GZ1357) and Guangxi Natural Science Foundation (2013GXNSFDA019012).
Fig. 1(A) Geographical distribution of amplified fragment length polymorphism (AFLP) genotypes of Gracilaria vermiculophylla along the coast of China. Insert (upper left) is the Unweighed Pair Group Method with Arithmetic mean (UPGMA) dendrogram inferred from AFLP. The approximate ranges of three marginal seas along the coast of China are also presented. (B) Barrier results of G. vermiculophylla along the coast of China. Spatial analysis of sites (n = 18) with two major barriers designated and labeled by red Roman numerals I and II. The line thickness corresponds to the segment distance among sampling sites. Insert (upper left) is a magnification view of the relationships among sites 7–18. YT, Yingtan Beach, Beihai, Guangxi; JH, Jinghaian, Beihai, Guangxi; TC, Techeng Island, Zhanjiang, Guangdong; NA, Nanao Island, Shantou, Guangdong; ZP, Zhangpu lvao, Zhangzhou, Fujian; SS, Shengsi Island, Zhoushan, Zhejiang; YY, Bathing Beach (No. 1), Qingdao, Shandong; EY, Bathing Beach (No. 2), Qingdao, Shandong; SY, Bathing Beach (No. 3), Qingdao, Shandong; YH, Yinghaiguoji, Qingdao, Shandong; LR, Shilaoren, Qingdao, Shandong; DC, Dongchu Island, Weihai, Shandong; JM, Jiming Island, Weihai, Shandong; SD, Shidao Island, Weihai, Shandong; CD, Changdao Island, Yantai, Shandong; HN, Huangnichuan, Dalian, Liaoning; HS, Heishijiao, Dalian, Liaoning; ZZ, Zhangzidao Island, Dalian, Liaoning. [Colour figure can be viewed at http://www.e-algae.org]. 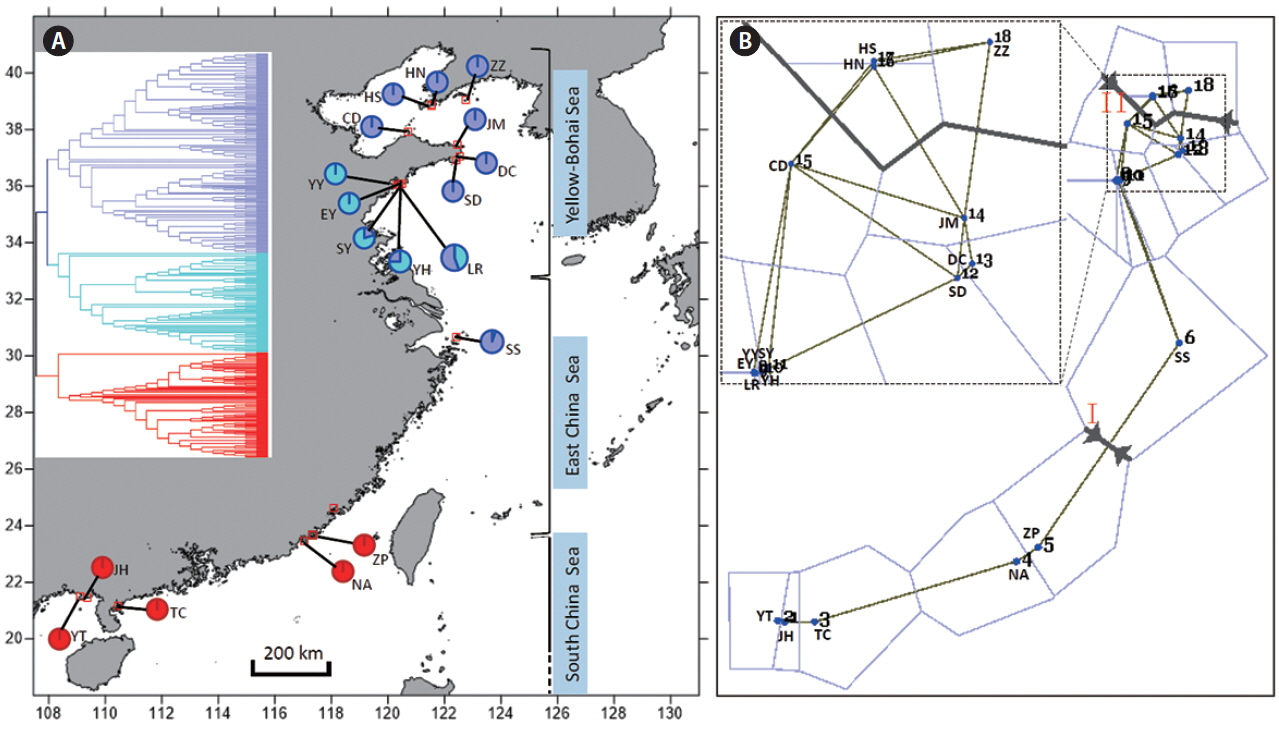
Fig. 2(A) Amplified fragment length polymorphism-based principal component analysis showed the relationships among the southern lineage and two subdivided northern clades. (B) Data output from the program STRUCTURE, runs averaged with CLUMPP and displayed with DISTRUCT. A genetic break was detected in Gracilaria vermiculophylla along the coast of China when K was set to 2, and a sub-genetic division was detected in the northern lineage when K was set to 3. [Colour figure can be viewed at http://www.e-algae.org]. 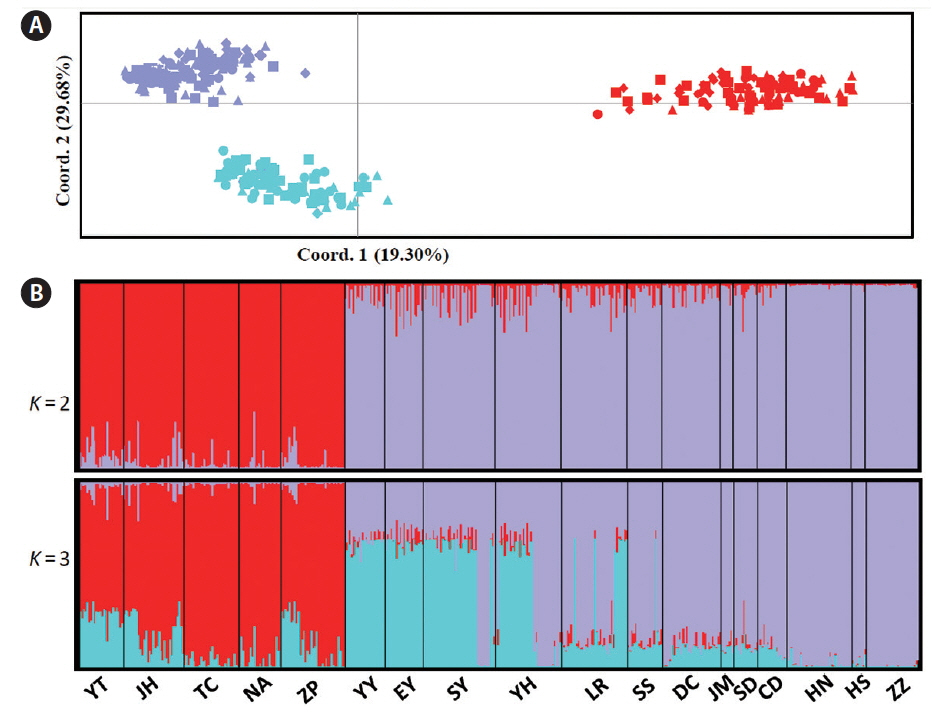
Table 1Genetic diversity of Gracilaria vermiculophylla along the coast of China inferred from AFLP
Table 2Pairwise Jost’ D genetic distances (lower-left diagonal) and FST estimates (upper-right diagonal) inferred from AFLP datasets
AFLP, amplified fragment length polymorphism; YT, Yingtan Beach, Beihai, Guangxi; JH, Jinghaian, Beihai, Guangxi; TC, Techeng Island, Zhanjiang, Guangdong; NA, Nanao Island, Shantou, Guangdong; ZP, Zhangpu lvao, Zhangzhou, Fujian; SS, Shengsi Island, Zhoushan, Zhejiang; YY, Bathing Beach (No. 1), Qingdao, Shandong; EY, Bathing Beach (No. 2), Qingdao, Shandong; SY, Bathing Beach (No. 3), Qingdao, Shandong; YH, Yinghaiguoji, Qingdao, Shandong; LR, Shilaoren, Qingdao, Shandong; DC, Dongchu Island, Weihai, Shandong; JM, Jiming Island, Weihai, Shandong; SD, Shidao Island, Weihai, Shandong; CD, Changdao Island, Yantai, Shandong; HN, Huangnichuan, Dalian, Liaoning; HS, Heishijiao, Dalian, Liaoning; ZZ, Zhangzidao Island, Dalian, Liaoning. REFERENCESAbreu, MH., Pereira, R., Yarish, C., Buschmann, AH. & Sousa-Pinto, I. 2011. IMTA with Gracilaria vermiculophylla: productivity and nutrient removal performance of the seaweed in a land-based pilot scale system. Aquaculture. 312:77–87.
Bonin, A., Bellemain, E., Bronken Eidesen, P., Pompanon, F., Brochmann, C. & Taberlet, P. 2004. How to track and assess genotyping errors in population genetic studies. Mol Ecol. 13:3261–3273.
Castric, V. & Bernatchez, L. 2003. The rise and fall of isolation by distance in the anadromous brook charr (Salvelinus fontinalis Mitchill). Genetics. 163:983–996.
Chao, A. & Shen, TJ. 2003. Program SPADE (Species Prediction and Diversity Estimation), Available from: http://chao.stat.nthu.edu.tw
. Accessed Apr 20, 2018
Cheang, CC., Chu, KH. & Ang, PO. 2010. Phylogeography of the marine macroalga Sargassum hemiphyllum (Phaeophyceae, Heterokontophyta) in northwestern Pacific. Mol Ecol. 19:2933–2948.
Diekmann, OE. & Serrão, EA. 2012. Range-edge genetic diversity: locally poor extant southern patches maintain a regionally diverse hotspot in the seagrass Zostera marina
. Mol Ecol. 21:1647–1657.
Doney, SC., Ruckelshaus, M., Duffy, JE., Barry, JP., Chan, F., English, CA., Galindo, HM., Grebmeier, JM., Hollowed, AB., Knowlton, N., Polovina, J., Rabalais, NN., Sydeman, WJ. & Talley, LD. 2012. Climate change impacts on marine ecosystems. Ann Rev Mar Sci. 4:11–37.
Eckert, CG., Samis, KE. & Lougheed, SC. 2008. Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol Ecol. 17:1170–1188.
Emerson, BC., Paradis, E. & Thébaud, C. 2001. Revealing the demographic histories of species using DNA sequences. Trends Ecol Evol. 16:707–716.
Excoffier, L. & Lischer, HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10:564–567.
Gibson, SY., van der Marel, RC. & Starzomski, BM. 2009. Climate change and conservation of leading-edge peripheral populations. Conserv Biol. 23:1369–1373.
Gorman, L., Kraemer, GP., Yarish, C., Boo, SM. & Kim, JK. 2017. The effects of temperature on the growth rate and nitrogen content of invasive Gracilaria vermiculophylla and native Gracilaria tikvahiae from Long Island Sound, USA. Algae. 32:57–66.
Hampe, A. & Petit, RJ. 2005. Conserving biodiversity under climate change: the rear edge matters. Ecol Lett. 8:461–467.
He, L., Zhang, A., Weese, D., Zhu, C., Jiang, C. & Qiao, Z. 2010. Late Pleistocene population expansion of Scylla paramamosain along the coast of China: a population dynamic response to the last interglacial sea level high stand. J Exp Mar Biol Ecol. 385:20–28.
Hu, Z-M. & Juan, L-B. 2014. Adaptation mechanisms and ecological consequences of seaweed invasions: a review case of agarophyte Gracilaria vermiculophylla
. Biol Invasions. 16:967–976.
Hu, Z-M., Kantachumpoo, A., Liu, R-Y., Sun, Z-M., Yao, J-T., Komatsu, T., Uwai, S. & Duan, D-L. 2018. A late Pleistocene marine glacial refugium in the south-west of Hainan Island, China: phylogeographical insights from the brown alga Sargassum polycystum
. J Biogeogr. 45:355–366.
Hu, Z-M., Li, J-J., Sun, Z-M., Gao, X., Yao, J-T., Choi, H-G., Endo, H. & Duan, D-L. 2017. Hidden diversity and phylogeographic history provide conservation insights for the edible seaweed Sargassum fusiforme in the Northwest Pacific. Evol Appl. 10:366–378.
Hu, Z-M., Li, J-J., Sun, Z-M., Oak, J-H., Zhang, J., Fresia, P., Grant, S. & Duan, D-L. 2015. Phylogeographic structure and deep lineage diversification of the red alga Chondrus ocellatus Holmes in the Northwest Pacific. Mol Ecol. 24:5020–5033.
Hu, Z-M., Uwai, S., Yu, S-H., Komatsu, T., Ajisaka, T. & Duan, D-L. 2011. Phylogeographic heterogeneity of the brown macroalga Sargassum horneri (Fucaceae) in the northwestern Pacific in relation to late Pleistocene glaciation and tectonic configurations. Mol Ecol. 20:3894–3909.
Jakobsson, M. & Rosenberg, NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 23:1801–1806.
Kim, SY., Weinberger, F. & Boo, SM. 2010. Genetic data hint at a common donor region for invasive Atlantic and Pacific populations of Gracilaria vermiculophylla (Gracilariales, Rhodophyta). J Phycol. 46:1346–1349.
Krueger-Hadfield, SA., Kollars, NM., Strand, AE., Byers, JE., Shainker, SJ., Terada, R., Greig, TW., Hammann, M., Murray, DC., Weinberger, F. & Sotka, EE. 2017. Genetic identification of source and likely vector of a widespread marine invader. Ecol Evol. 7:4432–4447.
Krueger-Hadfield, SA., Roze, D., Mauger, S. & Valero, M. 2013. Intergametophytic selfing and microgeographic genetic structure shape populations of the intertidal red seaweed Chondrus crispus
. Mol Ecol. 22:3242–3260.
Li, J-J., Hu, Z-M., Sun, Z-M., Yao, J-T., Liu, F-L., Fresia, P. & Duan, D-L. 2017. Historical isolation and contemporary gene flow drive population diversity of the brown alga Sargassum thunbergii along the coast of China. BMC Evol Biol. 17:246 pp.
Liu, RY., Sun, ZM., Yao, JT., Hu, ZM. & Duan, DL. 2016. Genetic diversity of the habitat-forming red alga Gracilaria vermiculophylla along the Chinese coasts. Biodivers Sci. 24:781–790, (in Chinese with English abstract).
Manni, F., Guérard, E. & Heyer, E. 2004. Goegraphic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier’s algorithm. Hum Biol. 76:173–190.
Neiva, J., Assis, J., Coelho, NC., Fernandes, F., Pearson, GA. & Serrão, EA. 2015. Genes left behind: climate change threatens cryptic genetic diversity in the canopy-forming seaweed Bifurcaria bifurcata
. PLoS ONE. 10:e0131530.
Neiva, J., Pearson, GA., Valero, M. & Serrão, EA. 2012. Drifting fronds and drifting alleles: range dynamics, local dispersal and habitat isolation shape the population structure of the estuarine seaweed Fucus ceranoides
. J Biogeogr. 39:1167–1178.
Nicastro, KR., Zardi, GI. & McQuaid, CD. 2010. Differential reproductive investment, attachment strength and mortality of invasive and indigenous mussels across heterogeneous environments. Biol Invasions. 12:2165–2177.
Olsen, JL., Stam, WT., Coyer, JA., Reusch, TBH., Billingham, M., Boström, C., Calvert, E., Christie, H., Granger, S., La Lumière, R., Milchakova, N., Oudot-Le Secq, M-P., Procaccini, G., Sanjabi, B., Serrão, E., Veldsink, J., Widdicombe, S. & Wyllie-Echeverria, S. 2004. North Atlantic phylogeography and large-scale population differentiation of the seagrass Zostera marina L. Mol Ecol. 13:1923–1941.
Pauls, SU., Nowak, C., Bálint, M. & Pfenninger, M. 2013. The impact of global climate change on genetic diversity within populations and species. Mol Ecol. 22:925–946.
Peakall, R. & Smouse, PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research: an update. Bioinformatics. 28:2537–2539.
Pritchard, JK., Stephens, M. & Donnelly, P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155:945–959.
Provan, J. & Maggs, CA. 2012. Unique genetic variation at a species’ rear edge is under threat from global climate change. Proc R Soc B Biol Sci. 279:39–47.
Ramírez, F., Afán, I., Davis, LS. & Chiaradia, A. 2017. Climate impacts on global hot spots of marine biodiversity. Sci Adv. 3:e1601198.
Ramus, AP., Silliman, BR., Thomsen, MS. & Long, ZT. 2017. An invasive foundation species enhances multifunctionality in a coastal ecosystem. Proc Natl Acad Sci U S A. 114:8580–8585.
Rosenberg, NA. 2004. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 4:137–138.
Sotka, EE., Baumgardner, AW., Bippus, PM., Destombe, C., Duermit, EA., Endo, H., Flanagan, BA., Kamiya, M., Lees, LE., Murren, CJ., Nakaoka, M., Shainker, SJ., Strand, AE., Terada, R., Valero, M., Weinberger, F. & Krueger-Hadfield, SA. 2018. Combining niche shift and population genetic analyses predicts rapid phenotypic evolution during invasion. Evol Appl. 11:781–793.
Terada, R. & Yamamoto, H. 2002. Review of Gracilaria vermiculophylla and other species in Japan and Asia. In : Abbott IA, Mcdermid KJ, editors Taxonomy of Economic Seaweeds with Reference to Some Pacific Species. 8:California Sea Grant College Program, University of California, La Jolla, CA, 215–224.
Tseng, CK. & Chen, SF. 1959. Reproductive biology and seeding development of Gracilaria asiatica in laboratory. Chin Sci Bull. 202–203, (in Chinese)..
Tseng, CK. & Xia, B-M. 1999. On the Gracilaria in the western Pacific and southeastern Asia region. Bot Mar. 42:209–218.
Vos, P., Hogers, R., Bleeker, M., Reijans, M., Van de Lee, T., Hornes, M., Friters, A., Pot, J., Paleman, J., Kuiper, M. & Zabeau, M. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acid Res. 23:4407–4414.
Wang, B-D., Wang, X-L. & Zhan, R. 2003. Nutrient conditions in the Yellow Sea and the East China Sea. Estuar Coast Shelf Sci. 58:127–136.
Wang, P-X. 1999. Response of western Pacific marginal seas to glacial cycles: paleoceanographic and sedimentological features. Mar Geol. 156:5–39.
Wernberg, T., Smale, DA., Tuya, F., Thomsen, MS., Langlois, TJ., de Bettignies, T., Bennett, S. & Rousseaux, CS. 2013. An extreme climatic event alters marine ecosystem structure in a global biodiversity hotspot. Nat Clim Change. 3:78–82.
Whitlock, R., Hipperson, H., Mannarelli, M., Butlin, RK. & Burke, T. 2008. An objective, rapid and reproducible method for scoring AFLP peak-height data that minimizes genotyping error. Mol Ecol Resour. 8:725–735.
Xu, SN., Wen, SS., Wu, WX. & He, PM. 2008. Bioremediation of caged fish aquaculture by the red alga Gracilaria verrucosa in an integrated multitrophic aquaculture system. Acta Ecol Sin. 28:1466–1475, (in Chinese with English abstract)..
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||